- Malware detection is a crucial aspect of cybersecurity.
- The objective of this study is to perform effective AI-powered malware detection and interpretation with the open-source H2O AutoML algorithm.
- H2O’s AutoML can be used for automating the Machine Learning (ML) workflow, which includes automatic training and tuning of many models within a user-specified time-limit. AutoML automates most of the steps in an ML pipeline, with a minimum amount of human effort and without compromising on its performance.
- The input Kaggle dataset is a result of a R&D about ML & Malware Detection. It was built using a Python Library and contains benign and malicious data from PE Files.
Table of Contents
Model Training
- Setting the working directory YOURPATH
import os
os.chdir('YOURPATH') # Set working directory
os. getcwd()
- Importing key libraries, reading the data and defining the H2O AutoML framework
import numpy as np
import pandas as pd
import pickle
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix, classification_report, accuracy_score
import h2o
from h2o.automl import H2OAutoML
h2o.init(max_mem_size='2G')
data = pd.read_csv('dataset_malwares.csv')
from h2o.frame import H2OFrame
data = H2OFrame(data)
- Splitting the data and training the model
train, test = data.split_frame(ratios=[0.6], seed=42)
from h2o.automl import H2OAutoML
automl = H2OAutoML(max_runtime_secs=600) # Set the maximum runtime in seconds
automl.train(x=data.columns[:-1], y='Malware', training_frame = train)
- Output logs:
AutoML: XGBoost is not available; skipping it.
Model Details
=============
H2OStackedEnsembleEstimator : Stacked Ensemble
Model Key: StackedEnsemble_AllModels_3_AutoML_2_20240211_162431
Model Summary for Stacked Ensemble:
key value
Stacking strategy cross_validation
Number of base models (used / total) 7/49
# GBM base models (used / total) 6/43
# DRF base models (used / total) 0/2
# GLM base models (used / total) 0/1
# DeepLearning base models (used / total) 1/3
Metalearner algorithm GLM
Metalearner fold assignment scheme Random
Metalearner nfolds 5
Metalearner fold_column None
Custom metalearner hyperparameters None
ModelMetricsRegressionGLM: stackedensemble
** Reported on train data. **
MSE: 0.0009636378361646813
RMSE: 0.031042516588779996
MAE: 0.008877334165576591
RMSLE: 0.02408848192111684
Mean Residual Deviance: 0.0009636378361646813
R^2: 0.9949665112607115
Null degrees of freedom: 9975
Residual degrees of freedom: 9968
Null deviance: 1909.8904021504356
Residual deviance: 9.61325105357886
AIC: -40952.613530641036
ModelMetricsRegressionGLM: stackedensemble
** Reported on cross-validation data. **
MSE: 0.00671905903384776
RMSE: 0.08196986662089771
MAE: 0.021946099162326017
RMSLE: 0.0578426755882866
Mean Residual Deviance: 0.00671905903384776
R^2: 0.9647434373633934
Null degrees of freedom: 11797
Residual degrees of freedom: 11791
Null deviance: 2249.4329600946585
Residual deviance: 79.27145848133587
AIC: -25525.84523210719
Cross-Validation Metrics Summary:
mean sd cv_1_valid cv_2_valid cv_3_valid cv_4_valid cv_5_valid
mae 0.0219382 0.0009372 0.0213027 0.0215373 0.0217661 0.0235886 0.0214964
mean_residual_deviance 0.0067135 0.0006209 0.0062748 0.006535 0.0069114 0.0076946 0.0061516
mse 0.0067135 0.0006209 0.0062748 0.006535 0.0069114 0.0076946 0.0061516
null_deviance 449.8866 16.695866 457.94696 466.87103 435.76636 460.2754 428.57324
r2 0.9647123 0.0034319 0.9682267 0.966678 0.9624521 0.9598925 0.9663121
residual_deviance 15.854292 1.6567098 14.526201 15.54023 16.33865 18.459446 14.406931
rmse 0.0818678 0.0037333 0.0792137 0.0808394 0.0831351 0.0877191 0.0784318
rmsle 0.0577558 0.0029666 0.0563536 0.0575922 0.0583191 0.0622814 0.0542329
- Performing test predictions of the automl leader
predictions = automl.leader.predict(test)
Model Optimization
- Selecting the best model from the leaderboard in terms of the ML evaluation metrics
from sklearn.metrics import precision_score, recall_score, f1_score
# Define evaluation metrics
def evaluate_model(true_labels, predicted_labels):
precision = precision_score(true_labels, predicted_labels)
recall = recall_score(true_labels, predicted_labels)
f1 = f1_score(true_labels, predicted_labels)
return precision, recall, f1
# List of models from the leaderboard
models = automl.leaderboard['model_id'].as_data_frame()['model_id'].tolist()
# Initialize best model
best_model = None
best_metrics = {'precision': 0, 'recall': 0, 'f1': 0}
# Evaluate each model and select the best one
for model_id in models:
# Make predictions
predictions = automl.leader.predict(test)
predicted_probabilities = predictions['predict'].as_data_frame()['predict'].values
predicted_labels = (predicted_probabilities > 0.5).astype(int)
# Evaluate the model
precision, recall, f1 = evaluate_model(true_labels, predicted_labels)
# Update best model if current model is better
if f1 > best_metrics['f1']:
best_model = model_id
best_metrics = {'precision': precision, 'recall': recall, 'f1': f1}
# Print information about the best model
print(f"Best Model: {best_model}")
print("Best Model Metrics:")
print(f"Precision: {best_metrics['precision']:.4f}")
print(f"Recall: {best_metrics['recall']:.4f}")
print(f"F1 Score: {best_metrics['f1']:.4f}")
- Output
Best Model: StackedEnsemble_AllModels_3_AutoML_2_20240211_162431
Best Model Metrics:
Precision: 0.9933
Recall: 0.9983
F1 Score: 0.9958
- Plotting the Pareto Front for H2O AutoML
pf = automl.pareto_front()
pf.figure()
pf
model_id rmse mse mae rmsle mean_residual_deviance training_time_ms predict_time_per_row_ms algo
StackedEnsemble_AllModels_3_AutoML_2_20240211_162431 0.0819699 0.00671906 0.0219461 0.0578427 0.00671906 510 0.151789 StackedEnsemble
StackedEnsemble_BestOfFamily_4_AutoML_2_20240211_162431 0.0842577 0.00709936 0.0224664 0.0593384 0.00709936 365 0.019229 StackedEnsemble
GBM_grid_1_AutoML_2_20240211_162431_model_29 0.0842626 0.00710019 0.0218079 0.0593146 0.00710019 3638 0.015837 GBM
GBM_grid_1_AutoML_2_20240211_162431_model_13 0.085033 0.00723061 0.0214841 0.0595172 0.00723061 1543 0.013421 GBM
GBM_grid_1_AutoML_2_20240211_162431_model_28 0.0859028 0.00737929 0.023693 0.0598147 0.00737929 1270 0.011117 GBM
GBM_grid_1_AutoML_2_20240211_162431_model_1 0.0872463 0.00761192 0.0176017 0.0605518 0.00761192 1716 0.009195 GBM
XRT_1_AutoML_2_20240211_162431 0.0988725 0.00977577 0.0295303 0.069559 0.00977577 972 0.006904 DRF
DRF_1_AutoML_2_20240211_162431 0.100821 0.0101649 0.02576 0.0709873 0.0101649 714 0.006876 DRF
GBM_grid_1_AutoML_2_20240211_162431_model_38 0.136559 0.0186483 0.0834828 0.101459 0.0186483 121 0.001727 GBM
GLM_1_AutoML_2_20240211_162431 0.253971 0.0645013 0.169792 nan 0.0645013 34 0.001054 GLM

Classification Report
- Extracting predicted probabilities and calculating the test accuracy
# Extract predicted probabilities
predicted_probabilities = predictions['predict'].as_data_frame()['predict'].values
# Convert predicted probabilities to class labels
predicted_labels = (predicted_probabilities > 0.5).astype(int)
# Ensure true_labels is of the same type (H2OFrame to Pandas DataFrame)
true_labels = test['Malware'].as_data_frame()['Malware'].values
# Calculate accuracy
accuracy = accuracy_score(true_labels, predicted_labels)
print(f'Test Accuracy: {accuracy * 100:.2f}%')
Test Accuracy: 99.37%
- Generating the ML classification report
# Confusion Matrix
cm = confusion_matrix(true_labels, predicted_labels)
print("Confusion Matrix:")
print(cm)
# Classification Report
report = classification_report(true_labels, predicted_labels)
print("Classification Report:")
print(report)
Confusion Matrix:
[[1950 39]
[ 10 5814]]
Classification Report:
precision recall f1-score support
0 0.99 0.98 0.99 1989
1 0.99 1.00 1.00 5824
accuracy 0.99 7813
macro avg 0.99 0.99 0.99 7813
weighted avg 0.99 0.99 0.99 7813
- Plotting the confusion matrix
# Assuming 'true_labels' and 'predicted_labels' are your true and predicted labels
cm = confusion_matrix(true_labels, predicted_labels)
# Plotting the Confusion Matrix
plt.figure(figsize=(7, 5))
sns.heatmap(cm, annot=True, fmt="d", cmap="Blues", xticklabels=['Non-Malware', 'Malware'], yticklabels=['Non-Malware', 'Malware'])
plt.title('Confusion Matrix')
plt.xlabel('Predicted Label')
plt.ylabel('True Label')
plt.show()

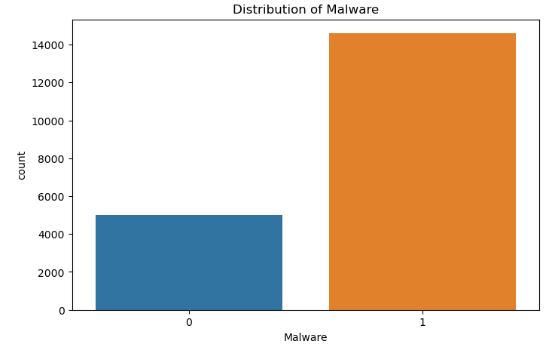
- Converting H2OFrame to Pandas DataFrame and plotting the Distribution of Malware
# Convert H2OFrame to Pandas DataFrame
data_pd = data.as_data_frame()
plt.figure(figsize=(8, 5))
sns.countplot(x='Malware', data=data_pd)
plt.title('Distribution of Malware')
plt.show()
- Plotting ROC and Precision-Recall Curves
from sklearn.metrics import roc_curve, precision_recall_curve, auc
# Assuming you have true_labels and predicted_probabilities
fpr, tpr, _ = roc_curve(true_labels, predicted_probabilities)
precision, recall, _ = precision_recall_curve(true_labels, predicted_probabilities)
plt.figure(figsize=(10, 5))
# ROC Curve
plt.subplot(1, 2, 1)
plt.plot(fpr, tpr, color='darkorange', lw=2, label='ROC curve (AUC = {:.2f})'.format(auc(fpr, tpr)))
plt.plot([0, 1], [0, 1], color='navy', lw=2, linestyle='--')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver Operating Characteristic (ROC) Curve')
plt.legend()
# Precision-Recall Curve
plt.subplot(1, 2, 2)
plt.plot(recall, precision, color='blue', lw=2, label='Precision-Recall curve')
plt.xlabel('Recall')
plt.ylabel('Precision')
plt.title('Precision-Recall Curve')
plt.legend()
plt.tight_layout()
plt.show()

Model Interpretations
- Leaderboard shows models with their metrics and their predictions for a given row. When provided with H2O AutoML object, the leaderboard shows 5-fold cross-validated metrics by default (depending on the H2O AutoML settings), otherwise it shows metrics computed on the frame. At most 20 models are shown by default.
model_id rmse mse mae rmsle mean_residual_deviance training_time_ms predict_time_per_row_ms algo predict
StackedEnsemble_AllModels_3_AutoML_2_20240211_162431 0.0773485 0.0059828 0.0200016 0.0530366 0.0059828 510 0.030086 StackedEnsemble 1.00686
GBM_grid_1_AutoML_2_20240211_162431_model_29 0.0783545 0.00613944 0.0192429 0.0535055 0.00613944 3638 0.016226 GBM 1.00446
StackedEnsemble_BestOfFamily_4_AutoML_2_20240211_162431 0.0784865 0.00616014 0.0198713 0.053641 0.00616014 365 0.016358 StackedEnsemble 1.00637
GBM_grid_1_AutoML_2_20240211_162431_model_13 0.0787041 0.00619433 0.0188041 0.05397 0.00619433 1543 0.012054 GBM 1.0046
StackedEnsemble_AllModels_2_AutoML_2_20240211_162431 0.0792631 0.00628264 0.0201192 0.0549249 0.00628264 592 0.04274 StackedEnsemble 1.00668
StackedEnsemble_AllModels_1_AutoML_2_20240211_162431 0.080609 0.00649781 0.0213402 0.0557984 0.00649781 613 0.049477 StackedEnsemble 1.00718
StackedEnsemble_BestOfFamily_3_AutoML_2_20240211_162431 0.0807443 0.00651964 0.0209621 0.0559776 0.00651964 298 0.014538 StackedEnsemble 1.00649
GBM_grid_1_AutoML_2_20240211_162431_model_16 0.0809532 0.00655342 0.0215635 0.0557101 0.00655342 4199 0.017187 GBM 1.00496
GBM_grid_1_AutoML_2_20240211_162431_model_19 0.0810464 0.00656851 0.0225983 0.0560315 0.00656851 1136 0.017246 GBM 1.00152
GBM_grid_1_AutoML_2_20240211_162431_model_35 0.0810495 0.00656902 0.0210852 0.0561155 0.00656902 990 0.014264 GBM 1.00752
[20 rows x 10 columns]
- SHAP explanation shows contribution of features for a given instance. The sum of the feature contributions and the bias term is equal to the raw prediction of the model, i.e., prediction before applying inverse link function. H2O implements TreeSHAP which when the features are correlated, can increase contribution of a feature that had no influence on the prediction.

- Partial dependence plot (PDP) gives a graphical depiction of the marginal effect of a variable on the response. The effect of a variable is measured in change in the mean response. PDP assumes independence between the feature for which is the PDP computed and the rest.
- Partial dependence plot for column=’MajorLinkerVersion’
column='MajorLinkerVersion'
pd_plot = automl.pd_multi_plot(test, column)

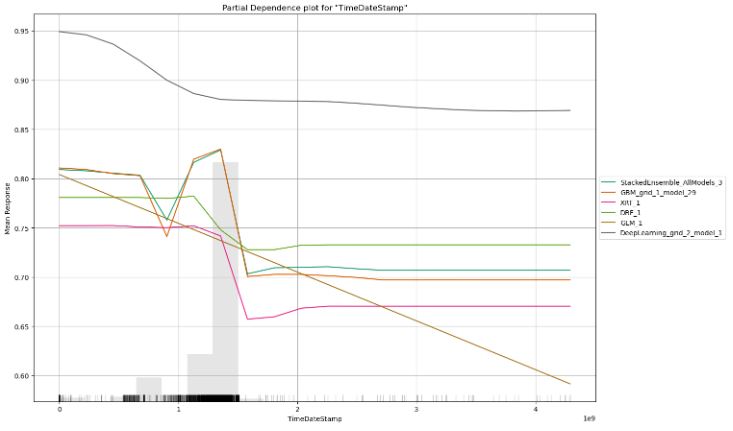
- Partial dependence plot for column=’TimeDateStamp’
column='TimeDateStamp'
pd_plot = automl.pd_multi_plot(test, column)
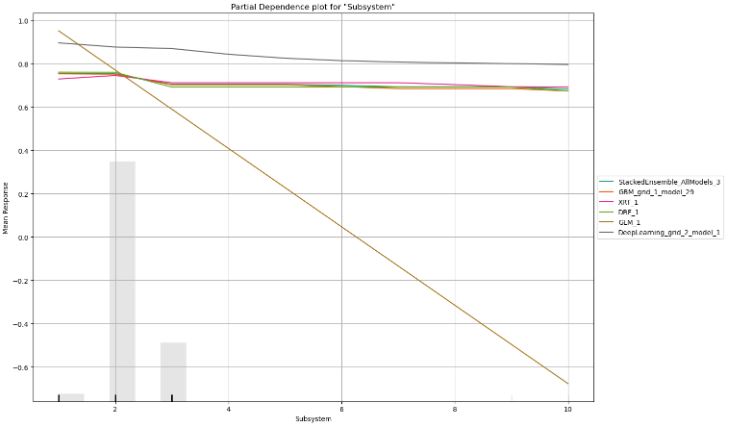
- Partial dependence plot for column=’Subsystem’
column='Subsystem'
pd_plot = automl.pd_multi_plot(test, column)
Summary
- Cyberattacks are currently the leading cause of concern in the tech world.
- The present proof-of-concept study is dedicated to evaluation and potential integration of ML algorithms to identify malware effectively and rapidly.
- Our validation tests confirm that the H2O AutoML workflow can accurately classify whether a given PE file is malware or not.
- We have used 7 GBM base models and 1 Deep Learning base model.
- Output Model Details: H2OStackedEnsembleEstimator or Stacked Ensemble (SE).
- Optimized SE model metrics are as follows:
- Precision: 0.9933
- Recall: 0.9983
- F1 Score: 0.9958
- SHAP explanation has shown contributions of features for a given instance.
- We have compared partial dependence plots for the top 3 features such as MajorLinkerVersion, TimeDateStamp, and Subsystem.
- This evaluation project supports related R&D products and solutions such as H2O’s AI and information security, Automatic Fraud Detection, and Auto_Malware_detection_with_h2o.
Explore More
- Robust Fake News Detection: NLP Algorithms for Deep Learning and Supervised ML in Python
- Comparison of 20 ML NLP Algorithms for SMS Spam-Ham Binary Classification
- Improved Multiple-Model ML/DL Credit Card Fraud Detection: F1=88% & ROC=91%
- Data-Driven ML Credit Card Fraud Detection
- Bank Note Authentication – Classification
- Cybersecurity Monthly Update
- Cybersecurity Round-Up
- Technology Focus Weekly Update 16 Oct ’22
- Cloud-Native Tech Autumn 2022 Fair
One-Time
Monthly
Yearly
Make a one-time donation
Make a monthly donation
Make a yearly donation
Choose an amount
€5.00
€15.00
€100.00
€5.00
€15.00
€100.00
€5.00
€15.00
€100.00
Or enter a custom amount
€
Your contribution is appreciated.
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthlyDonate yearly

Leave a reply to H2O AutoML Malware Detection F1 0.99 by Optimizing 8 ML Models with SHAP XAI | by Alexzap | Jul, 2024 – Artificial Intelligence Article Cancel reply