- Machine learning (ML) is an important decision support tool for crop yield prediction, including supporting decisions on what crops to grow and what to do during the growing season of the crops.
- Following recent research studies, this case example investigates whether coupling crop modeling, exploratory data analysis (EDA), ML yield prediction, recommendation, and multi-label classification algorithms yield competitive results with respect to the crop growth model. The main objectives are to explore whether a hybrid approach would result in better predictions, investigate which combinations of hybrid models provide the most accurate predictions, and determine the features from the crop modeling that are most effective to be integrated with ML for smart farming.
Table of Contents
- CatBoost/SHAP: Indian Crop Yield
- ML-Powered Indian Agriculture
- Global Crop Yield Prediction
- Crop Recommendation
- Effects of Climate Changes
- Summary
- Explore More
CatBoost/SHAP: Indian Crop Yield
- We begin our study with the Agricultural crop yield prediction in Indian states using the CatBoost & SHAP algorithms in Python.
- Agricultural Crop Yield in Indian States Dataset (DAT1):
This dataset encompasses agricultural data for multiple crops cultivated across various states in India from the year 1997 till 2020. The dataset provides crucial features related to crop yield prediction, including crop types, crop years, cropping seasons, states, areas under cultivation, production quantities, annual rainfall, fertilizer usage, pesticide usage, and calculated yields.
- Data Columns Description:
- Crop: The name of the crop cultivated.
- Crop_Year: The year in which the crop was grown.
- Season: The specific cropping season (e.g., Kharif, Rabi, Whole Year).
- State: The Indian state where the crop was cultivated.
- Area: The total land area (in hectares) under cultivation for the specific crop.
- Production: The quantity of crop production (in metric tons).
- Annual_Rainfall: The annual rainfall received in the crop-growing region (in mm).
- Fertilizer: The total amount of fertilizer used for the crop (in kilograms).
- Pesticide: The total amount of pesticide used for the crop (in kilograms).
- Yield: The calculated crop yield (production per unit area).
- Set the working directory YOURPATH
import os
os.chdir('YOURPATH')
os. getcwd()
- Import and install the key libraries
!pip install feature_engine
# import libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import warnings
warnings.filterwarnings("ignore")
import shap
import matplotlib.pyplot as plt
from catboost import Pool, CatBoostRegressor
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
from feature_engine.encoding import RareLabelEncoder
from sklearn.feature_extraction.text import CountVectorizer
import ast
- Read the input Kaggle dataset DAT1
df = pd.read_csv("crop_yield.csv").drop_duplicates()
print(df.shape)
df.sample(5).T
(19689, 10)

df.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 19689 entries, 0 to 19688
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Crop 19689 non-null object
1 Crop_Year 19689 non-null int64
2 Season 19689 non-null object
3 State 19689 non-null object
4 Area 19689 non-null float64
5 Production 19689 non-null int64
6 Annual_Rainfall 19689 non-null float64
7 Fertilizer 19689 non-null float64
8 Pesticide 19689 non-null float64
9 Yield 19689 non-null float64
dtypes: float64(5), int64(2), object(3)
memory usage: 1.7+ MB
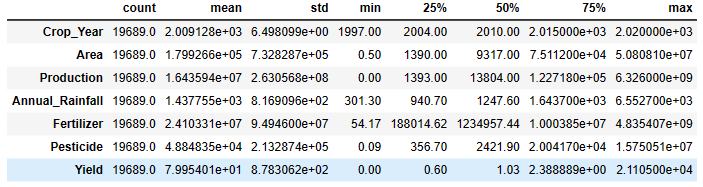
df.describe().T
- EDA via SweetViz
import sweetviz as sv
import pandas as pd
# Create an analysis report for the data
report = sv.analyze(df)
# Display the report
report.show_html()
Report SWEETVIZ_REPORT.html was generated! NOTEBOOK/COLAB USERS: the web browser MAY not pop up, regardless, the report IS saved in your notebook/colab files.

SweetViz associations:
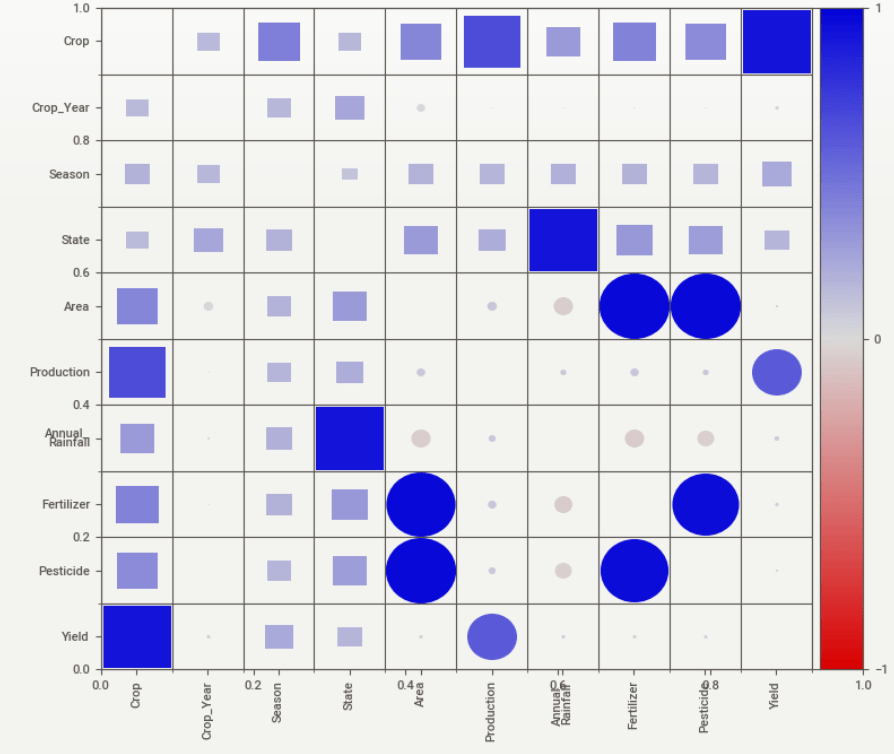
Here, Squares are categorical associations (uncertainty coefficient & correlation ratio) from 0 to 1. The uncertainty coefficient is asymmetrical (i.e. ROW LABEL values indicate how much they PROVIDE INFORMATION to each LABEL at the TOP). Circles are the symmetrical numerical correlations (Pearson’s) from -1 to 1. The trivial diagonal is intentionally left blank for clarity.
- Data preparation
# select records with non-zero yield
df = df[df['Yield']>0]
# select main label
main_label = 'log10_yield'
df[main_label] = df['Yield'].apply(lambda x: np.log10(x))
# divide fertilizer and pesticide by area (in kg/hectare) and annual rainfall (in mm), log10-transform and group into larger bins
for col in ['Fertilizer', 'Pesticide']:
df[f'log10_{col}_Area'] = (df[col]/df['Area']).apply(lambda x: 1/10*round(10*np.log10(x)))
df['log10_Annual_Rainfall'] = df['Annual_Rainfall'].apply(lambda x: 1/10*round(10*np.log10(x)))
# set up the rare label encoder limiting number of categories to max_n_categories
for col in ['Crop', 'Season', 'State']:
encoder = RareLabelEncoder(n_categories=1, max_n_categories=50, replace_with='Other', tol=25/df.shape[0])
df[col] = encoder.fit_transform(df[[col]])
# drop unused columns
cols2drop = ['Production', 'Yield', 'Fertilizer', 'Pesticide', 'Area', 'Annual_Rainfall']
df = df.drop(cols2drop, axis=1)
- Train/test data splitting as 70:30
y = df[main_label].values.reshape(-1,)
X = df.drop([main_label], axis=1)
cat_cols = df.select_dtypes(include=['object']).columns
cat_cols_idx = [list(X.columns).index(c) for c in cat_cols]
X_train,X_test,y_train,y_test = train_test_split(X, y, test_size=0.3, random_state=0)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
- Model training, validation and prediction
# initialize Pool
train_pool = Pool(X_train,
y_train,
cat_features=cat_cols_idx)
test_pool = Pool(X_test,
y_test,
cat_features=cat_cols_idx)
# specify the training parameters
model = CatBoostRegressor(iterations=1000,
depth=5,
verbose=0,
learning_rate=0.15,
loss_function='RMSE')
# train the model
model.fit(train_pool)
# make the prediction using the resulting model
y_train_pred = model.predict(train_pool)
y_test_pred = model.predict(test_pool)
rmse_train = mean_squared_error(y_train, y_train_pred, squared=False)
rmse_test = mean_squared_error(y_test, y_test_pred, squared=False)
print(f"RMSE score for train {round(rmse_train,3)} dex, and for test {round(rmse_test,3)} dex")
RMSE score for train 0.152 dex, and for test 0.168 dex
# Baseline scores (assuming the same prediction for all data samples)
rmse_bs_train = mean_squared_error(y_train, [np.mean(y_train)]*len(y_train), squared=False)
rmse_bs_test = mean_squared_error(y_test, [np.mean(y_train)]*len(y_test), squared=False)
print(f"RMSE baseline score for train {round(rmse_bs_train,3)} dex, and for test {round(rmse_bs_test,3)} dex")
RMSE baseline score for train 0.648 dex, and for test 0.662 dex
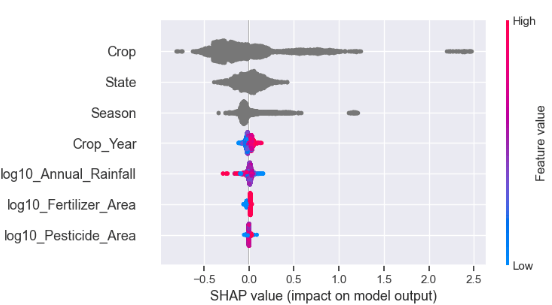
- SHAP initialization and summary value plot
shap.initjs()
ex = shap.TreeExplainer(model)
shap_values = ex.shap_values(X_test)
shap.summary_plot(shap_values, X_test)
- Expected vs actual log10 yield values
expected_values = ex.expected_value
print(f"Average predicted log10 yield is {round(expected_values,3)} dex")
print(f"Average actual log10 yield is {round(np.mean(y_test),3)} dex")
Average predicted log10 yield is 0.166 dex
Average actual log10 yield is 0.166 dex
- SHAP values for log10 columns vs log10 yield
def show_shap(col, shap_values=shap_values, label=main_label, X_test=X_test, ylabel='dex'):
df_infl = X_test.copy()
df_infl['shap_'] = shap_values[:,df_infl.columns.tolist().index(col)]
gain = round(df_infl.groupby(col).mean()['shap_'],4)
gain_std = round(df_infl.groupby(col).std()['shap_'],4)
cnt = df_infl.groupby(col).count()['shap_']
dd_dict = {'col': list(gain.index), 'gain': list(gain.values), 'gain_std': list(gain_std.values), 'count': cnt}
df_res = pd.DataFrame.from_dict(dd_dict).sort_values('gain', ascending=False).set_index('col')
plt.figure(figsize=(12,5))
plt.errorbar(df_res.index, df_res['gain'], yerr=df_res['gain_std'], fmt="o", color="r")
plt.title(f'SHAP values for column {col}, label {label}')
plt.ylabel(ylabel)
plt.tick_params(axis="x", rotation=90)
plt.show();
print(df_res)
return
for col in X_test.columns:
print()
print(col)
print()
show_shap(col, shap_values, label=main_label, X_test=X_test)
Crop
gain gain_std count
col
Coconut 2.3294 0.1996 55
Sugarcane 0.9245 0.1310 185
Tapioca 0.8204 0.0767 61
Jute 0.6929 0.1759 46
Banana 0.6839 0.1331 68
Potato 0.6438 0.0909 178
Onion 0.5671 0.1460 143
Mesta 0.5604 0.1945 49
Sweet potato 0.4691 0.0912 72
Ginger 0.4002 0.1407 97
Garlic 0.1264 0.0891 76
Wheat 0.1169 0.0587 153
Maize 0.1059 0.0672 311
Rice 0.0945 0.0729 384
Barley -0.0268 0.0749 82
Cotton(lint) -0.0511 0.0750 146
Turmeric -0.0662 0.1038 105
Groundnut -0.0683 0.0526 217
Bajra -0.0751 0.1232 170
Arecanut -0.0916 0.1117 42
Ragi -0.1053 0.0804 150
Peas & beans (Pulses) -0.1158 0.0847 115
Dry chillies -0.1196 0.1651 113
Soyabean -0.1201 0.0592 121
Jowar -0.1299 0.0718 136
Tobacco -0.1349 0.1737 94
Sunflower -0.1842 0.0569 131
Arhar/Tur -0.1927 0.0588 158
Gram -0.2001 0.0586 141
Other Cereals -0.2332 0.0541 56
Other Rabi pulses -0.2515 0.0787 102
Small millets -0.2562 0.0500 144
Cowpea(Lobia) -0.2679 0.0893 36
Other Kharif pulses -0.2808 0.0557 112
other oilseeds -0.2836 0.1094 42
Castor seed -0.2889 0.0858 80
Masoor -0.2919 0.0530 90
Rapeseed &Mustard -0.2971 0.0954 146
Safflower -0.3099 0.0389 56
Sannhamp -0.3295 0.2001 43
Linseed -0.3501 0.0361 86
Niger seed -0.3542 0.0463 59
Horse-gram -0.3634 0.0543 116
Urad -0.3636 0.0596 248
Moong(Green Gram) -0.3797 0.0658 203
Cashewnut -0.3963 0.1056 30
Other -0.4002 0.2008 79
Black pepper -0.4069 0.1651 37
Coriander -0.4146 0.1051 57
Moth -0.4195 0.0815 32
Sesamum -0.4215 0.0527 221
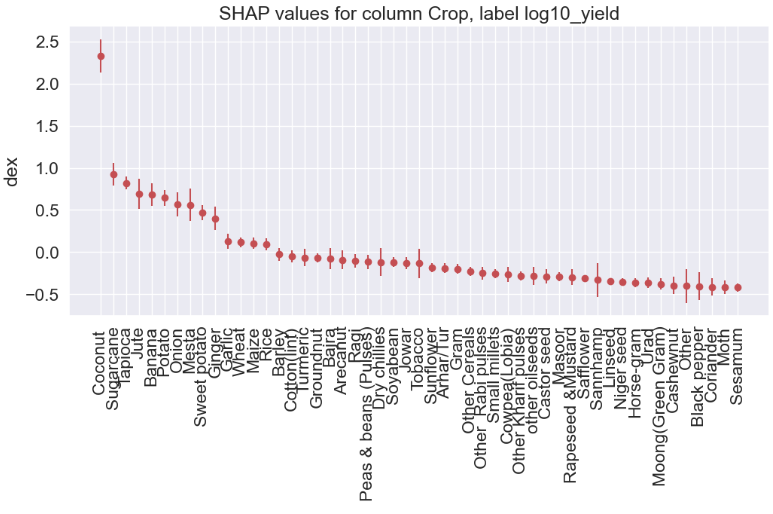
Crop_Year
gain gain_std count
col
2020 0.0488 0.0102 15
2019 0.0422 0.0205 321
2018 0.0407 0.0180 319
2017 0.0377 0.0200 309
2013 0.0268 0.0109 289
2014 0.0267 0.0131 264
2016 0.0252 0.0175 316
2012 0.0250 0.0142 262
2015 0.0238 0.0144 297
2011 -0.0000 0.0125 288
2010 -0.0036 0.0092 269
2007 -0.0092 0.0103 221
2008 -0.0114 0.0091 259
2006 -0.0148 0.0110 262
2009 -0.0153 0.0107 245
2005 -0.0173 0.0115 233
2004 -0.0175 0.0105 232
2003 -0.0177 0.0122 260
1999 -0.0248 0.0214 203
2001 -0.0261 0.0134 236
2002 -0.0308 0.0153 237
2000 -0.0316 0.0187 239
1998 -0.0376 0.0233 177
1997 -0.0511 0.0287 121
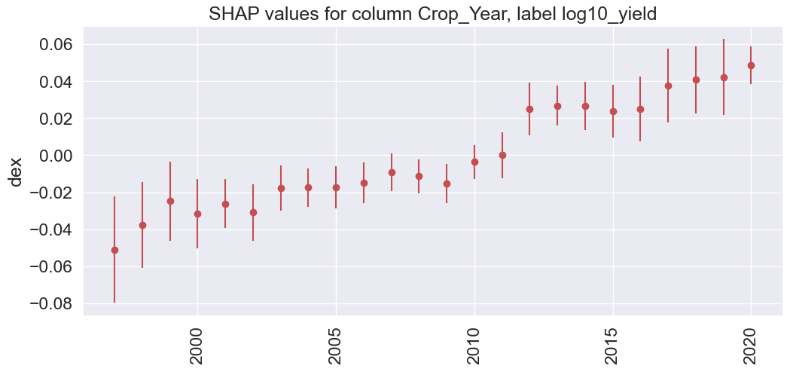
Season
gain gain_std count
col
Whole Year 0.2325 0.2568 1078
Summer 0.0551 0.0395 369
Winter 0.0032 0.0770 124
Rabi -0.0439 0.0326 1645
Kharif -0.0642 0.0427 2542
Autumn -0.0786 0.0443 116
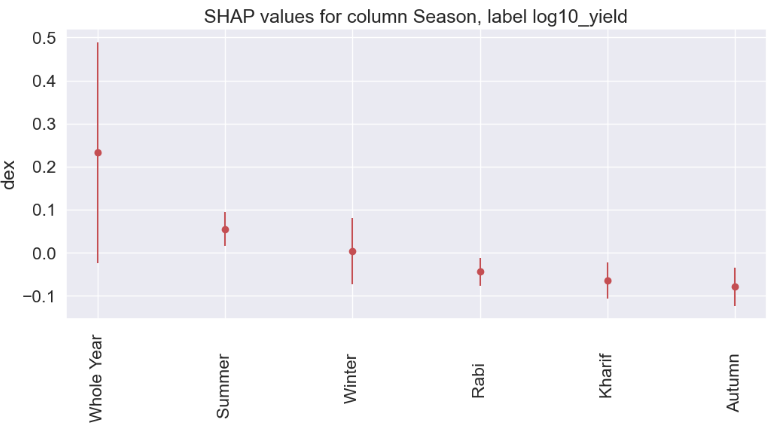
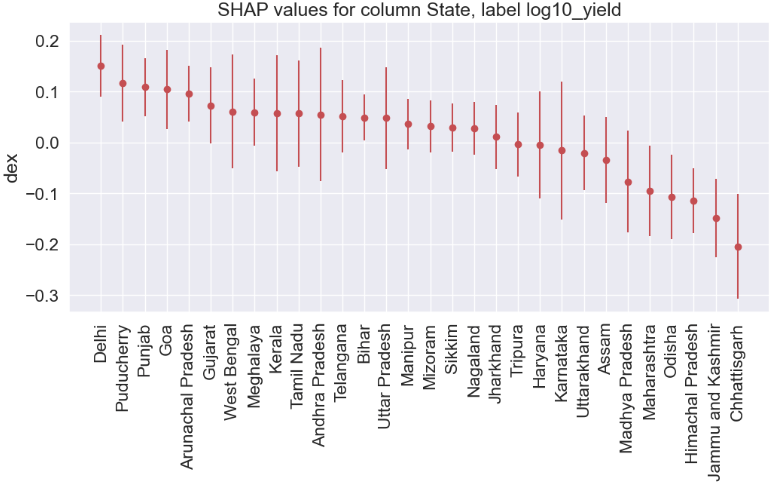
State
gain gain_std count
col
Delhi 0.1506 0.0605 54
Puducherry 0.1170 0.0756 208
Punjab 0.1086 0.0567 129
Goa 0.1046 0.0778 68
Arunachal Pradesh 0.0961 0.0548 82
Gujarat 0.0729 0.0755 256
West Bengal 0.0610 0.1118 354
Meghalaya 0.0595 0.0662 175
Kerala 0.0579 0.1140 154
Tamil Nadu 0.0567 0.1042 237
Andhra Pradesh 0.0547 0.1312 382
Telangana 0.0514 0.0705 115
Bihar 0.0492 0.0456 263
Uttar Pradesh 0.0478 0.1003 220
Manipur 0.0360 0.0498 123
Mizoram 0.0317 0.0508 135
Sikkim 0.0291 0.0479 85
Nagaland 0.0282 0.0517 224
Jharkhand 0.0108 0.0624 82
Tripura -0.0036 0.0627 144
Haryana -0.0049 0.1052 163
Karnataka -0.0156 0.1353 415
Uttarakhand -0.0206 0.0734 231
Assam -0.0346 0.0840 214
Madhya Pradesh -0.0773 0.1000 249
Maharashtra -0.0953 0.0888 228
Odisha -0.1070 0.0836 238
Himachal Pradesh -0.1144 0.0643 181
Jammu and Kashmir -0.1489 0.0770 187
Chhattisgarh -0.2043 0.1029 278
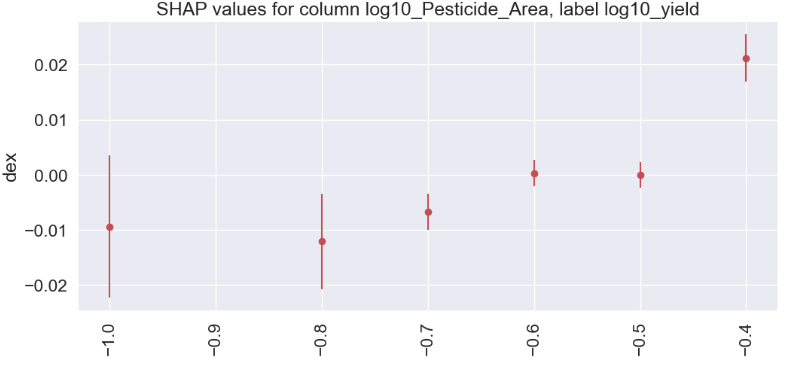
log10_Pesticide_Area
gain gain_std count
col
-0.4 0.0212 0.0043 645
-0.6 0.0003 0.0024 1733
-0.5 0.0000 0.0023 2044
-0.7 -0.0067 0.0033 727
-1.0 -0.0094 0.0129 259
-0.8 -0.0121 0.0087 466
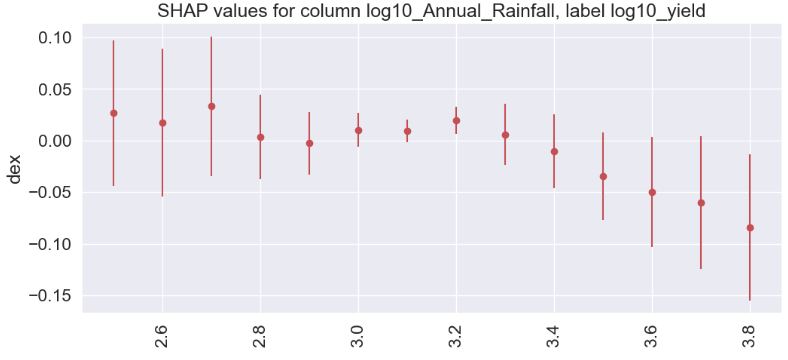
- log10_Annual_Rainfall
gain gain_std count
col
2.7 0.0332 0.0678 143
2.5 0.0267 0.0705 72
3.2 0.0196 0.0134 978
2.6 0.0174 0.0719 88
3.0 0.0104 0.0166 1175
3.1 0.0093 0.0111 1334
3.3 0.0058 0.0296 464
2.8 0.0036 0.0407 293
2.9 -0.0027 0.0302 554
3.4 -0.0105 0.0359 394
3.5 -0.0346 0.0426 241
3.6 -0.0499 0.0532 87
3.7 -0.0599 0.0642 23
3.8 -0.0840 0.0708 28
ML-Powered Indian Agriculture
- In this section, we will focus on the Indian Agricultural Crop Yield Predictions using supervised ML algorithms and the aforementioned input dataset DAT1. Our objective is to overcome observed shortcomings of basic linear regression applied to DAT1.
- Basic imports and load input dataset DAT1
# Importing Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import warnings
warnings.filterwarnings('ignore')
# Loading the dataset
df = pd.read_csv('crop_yield.csv')
- Exploratory Data Analysis (EDA)
Measure of Yield over the year < 2020
df_year = df[df['Crop_Year']!=2020]
year_yield = df_year.groupby('Crop_Year').sum()
plt.figure(figsize = (12,5))
plt.plot(year_yield.index, year_yield['Yield'],color='blue', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='yellow')
plt.xlabel('Year')
plt.ylabel('Yield')
plt.title('Measure of Yield over the year')
plt.show()

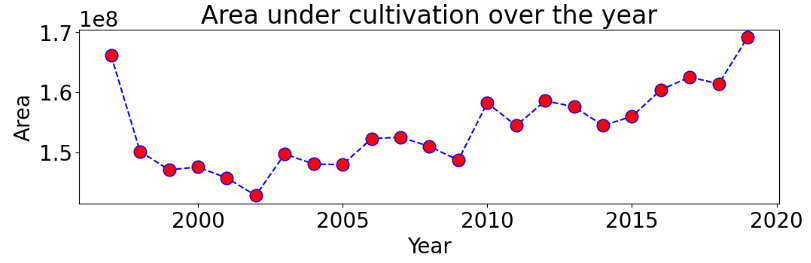
Area under cultivation over the year
plt.figure(figsize = (12,3))
plt.plot(year_yield.index, year_yield['Area'],color='blue', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='red')
plt.xlabel('Year')
plt.ylabel('Area')
plt.title('Area under cultivation over the year')
plt.show()
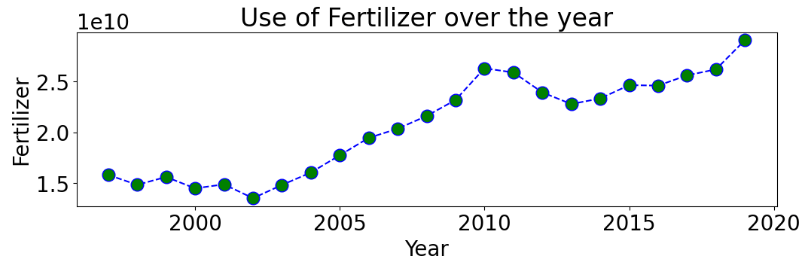
Use of Fertilizer over the year
plt.figure(figsize = (12,3))
plt.plot(year_yield.index, year_yield['Fertilizer'],color='blue', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='green')
plt.xlabel('Year')
plt.ylabel('Fertilizer')
plt.title('Use of Fertilizer over the year')
plt.show()
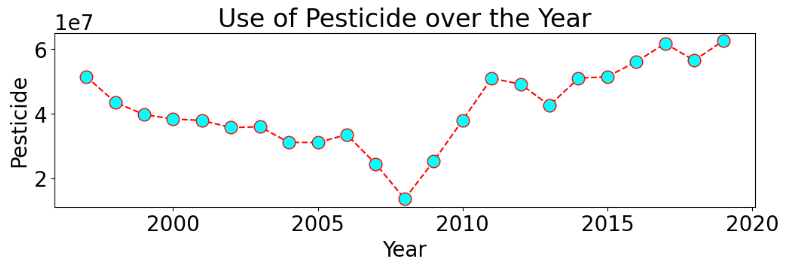
Use of Pesticide over the Year
plt.figure(figsize = (12,3))
plt.plot(year_yield.index, year_yield['Pesticide'],color='red', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='cyan')
plt.xlabel('Year')
plt.ylabel('Pesticide')
plt.title('Use of Pesticide over the Year')
plt.show()
Yield vs Region/State
df_state = df.groupby('State').sum()
df_state.sort_values(by = 'Yield', inplace=True, ascending = False)
df_state['Region'] = ['States' for i in range(len(df_state))]
fig = px.bar(df_state, x='Region', y = 'Yield', color=df_state.index, hover_data=['Yield'])
fig.show()

Bar Plot Annual Rainfall vs State
plt.figure(figsize = (15,8))
sns.barplot(x = df_state.index, y=df_state['Annual_Rainfall'], palette = 'gnuplot')
plt.xticks(rotation = 90)
plt.savefig('annualrainfallstate.png')

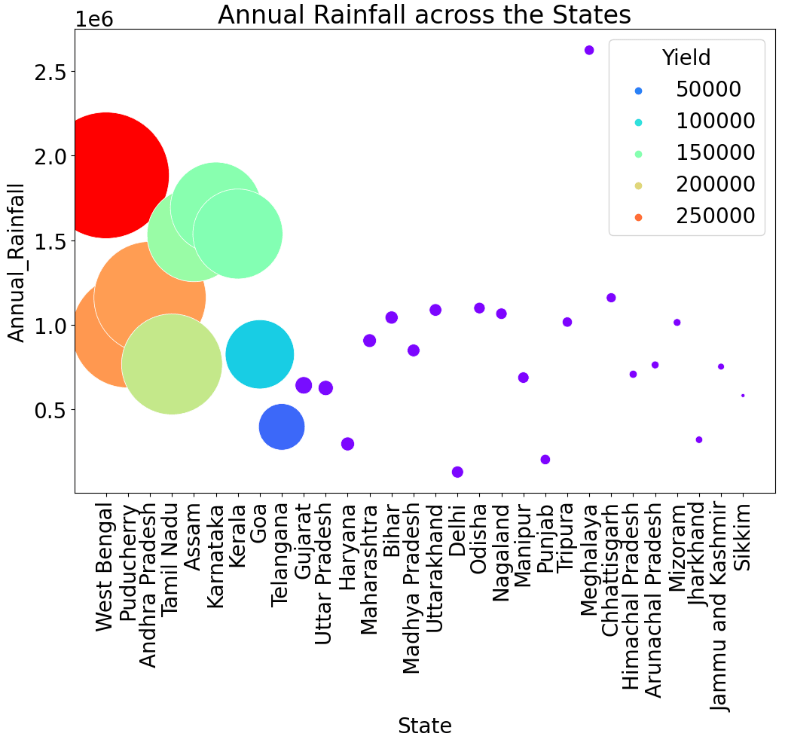
Bubble Plot Annual Rainfall and Yield across the States
plt.figure(figsize=(12,8))
sns.scatterplot(x=df_state.index, y = df_state['Annual_Rainfall'], palette='rainbow', hue = df_state['Yield'],s=df_state['Yield']/20)
plt.xticks(rotation=90)
plt.title('Annual Rainfall across the States')
plt.show()
Bubble Plot Fertilizer and Yield vs State
plt.figure(figsize=(12,8))
sns.scatterplot(x=df_state.index, y=df_state['Fertilizer'], palette='spring', hue = df_state['Yield'],s=df_state['Yield']/20)
plt.xticks(rotation=90)
plt.title('Use of Fertilizer in Different States')
plt.show()

Plotly Bar Plot Area vs Season
df_Seas = df[df['Season']!='Whole Year ']
df_season = df_Seas.groupby('Season').sum()
fig = px.bar(df_season, y = 'Area', color=df_season.index, hover_data=['Area'],text = 'Area')
fig.update_layout(
font=dict(
family="Courier New, monospace",
size=18, # Set the font size here
color="RebeccaPurple"
)
)
fig.show()

Plotly Pie Chart Yield vs Season
fig = px.sunburst(df_season, path=[df_season.index, 'Yield'], values='Yield',
color=df_season.index, hover_data=['Yield'])
fig.update_layout(
font=dict(
family="Courier New, monospace",
size=18, # Set the font size here
color="RebeccaPurple"
))
fig.show()

States and Crops where Yield=0
df_yz = df[df['Yield']==0]
plt.figure(figsize = (25,15))
sns.catplot(y="State", x="Crop",data=df_yz, aspect = 3, palette ='inferno')
plt.xticks(rotation=90)
plt.title('States and Crops where Yield=0')
plt.show()

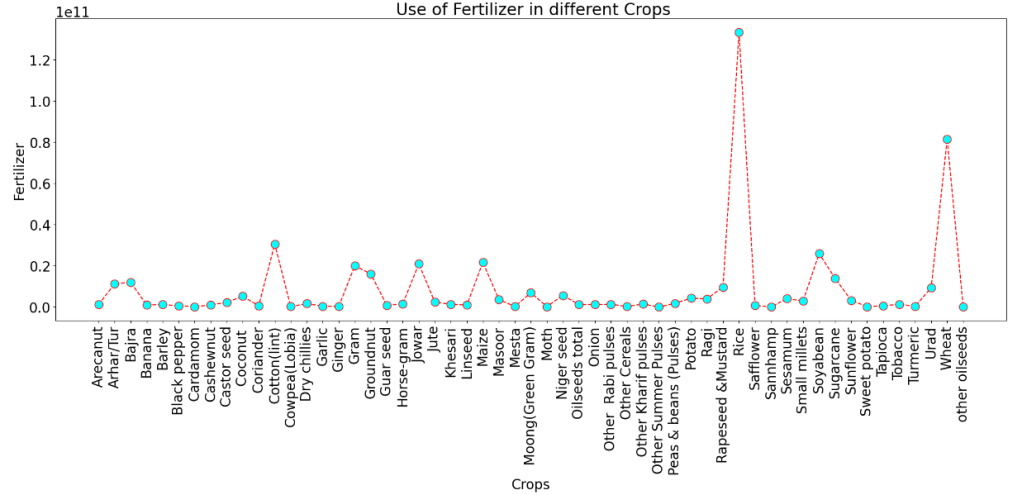
Use of Fertilizer in different Crops for States with Yield>0
df_ynz = df[df['Yield']>0] # where yield is more than zero
df_crop = df_ynz.groupby('Crop').sum()
plt.figure(figsize = (25,8))
plt.plot(df_crop.index, df_crop['Fertilizer'],color='red', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='cyan')
plt.rcParams.update({'font.size': 20})
plt.xlabel('Crops')
plt.ylabel('Fertilizer')
plt.title(' Use of Fertilizer in different Crops')
plt.xticks(rotation=90)
plt.show()
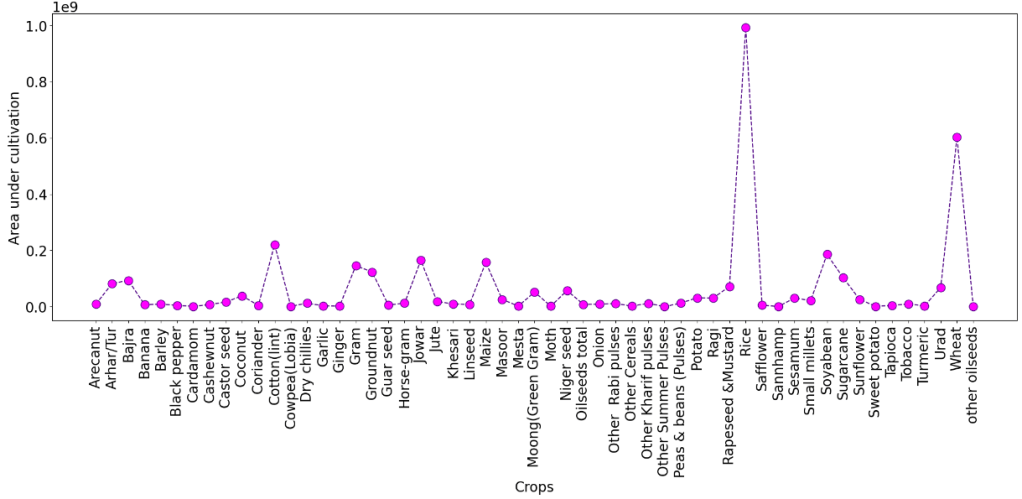
Area under Cultivation in different Crops for States with Yield>0
plt.figure(figsize = (25,8))
plt.rcParams.update({'font.size': 20})
plt.plot(df_crop.index, df_crop['Area'],color='indigo', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='fuchsia')
plt.xlabel('Crops')
plt.ylabel('Area under cultivation')
plt.xticks(rotation=90)
plt.show()
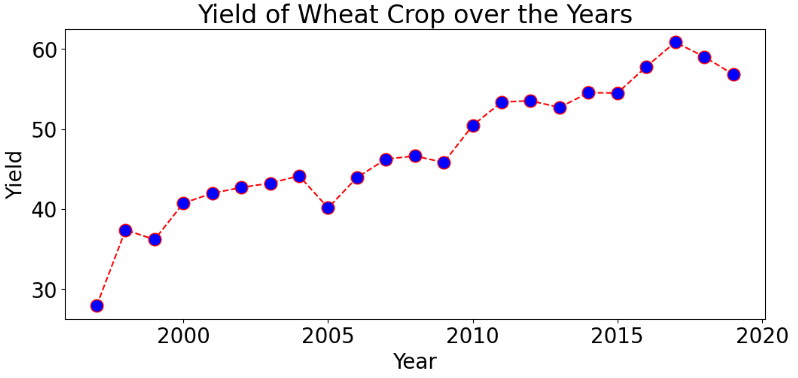
Yield of Wheat Crop over the Years
df_wheat = df[df['Crop']=='Wheat']
df_wheat.reset_index(drop=True,inplace=True)
df_wheat1 = df_wheat[df_wheat['Crop_Year']!=2020]
df_wheat_year = df_wheat1.groupby('Crop_Year').sum()
plt.figure(figsize = (12,5))
plt.plot(df_wheat_year.index, df_wheat_year['Yield'],color='red', linestyle='dashed', marker='o',
markersize=12, markerfacecolor='blue')
plt.xlabel('Year')
plt.ylabel('Yield')
plt.title('Yield of Wheat Crop over the Years')
plt.show()
Empirical Probability Analysis, Train/Test Split and Power Transformation
df1 = df.copy()
df1 = df1.drop(['Crop_Year','Pesticide'], axis = 1)
category_columns = df1.select_dtypes(include = ['object']).columns
df1 = pd.get_dummies(df1, columns = category_columns, drop_first=True)
x = df1.drop(['Yield'], axis = 1)
y = df1[['Yield']]
#split the data into training and test set as 70:30 %
from sklearn.model_selection import train_test_split
x_train, x_test, y_train,y_test = train_test_split(x, y, test_size = 0.3, random_state = 42)
from sklearn.preprocessing import PowerTransformer
pt = PowerTransformer(method='yeo-johnson')
x_train_transform1 = pt.fit_transform(x_train)
x_test_transform1 = pt.fit_transform(x_test)
df_trans = pd.DataFrame(x_train_transform1, columns=x_train.columns)
Histograms of transformed data: Area, Production, Annual Rainfall, and Fertilizer
plt.figure(figsize=(15,20))
plt.subplot(4,2,1)
sns.distplot(df_trans['Area'],bins = 20,color = 'red')
plt.subplot(4,2,2)
sns.distplot(df_trans['Production'],bins = 10,color = 'green')
plt.subplot(4,2,3)
sns.distplot(df_trans['Annual_Rainfall'],bins = 10,color = 'fuchsia')
plt.subplot(4,2,4)
sns.distplot(df_trans['Fertilizer'],bins = 10, color = 'indigo')
plt.show()

Q-Q probability plots of transformed data: Area, Production, Annual Rainfall, and Fertilizer
plt.figure(figsize=(15,40))
plt.subplot(4,2,1)
stats.probplot(df_trans['Area'], dist = 'norm', plot = plt)
plt.subplot(4,2,2)
stats.probplot(df_trans['Production'], dist = 'norm', plot = plt)
plt.subplot(4,2,3)
stats.probplot(df_trans['Annual_Rainfall'], dist = 'norm', plot = plt)
plt.subplot(4,2,4)
stats.probplot(df_trans['Fertilizer'], dist = 'norm', plot = plt)
plt.show()
Q-Q Plot Area Q-Q Plot Production
Q-Q Plot Annual Rainfall Q-Q Plot Fertilizer
- Linear Regression before Transform
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
lr = LinearRegression()
lr.fit(x_train,y_train)
y_pred_train = lr.predict(x_train)
print("Training Accuracy : ",r2_score(y_train,y_pred_train))
y_pred_test = lr.predict(x_test)
print("Test Accuracy : ",r2_score(y_test,y_pred_test))
Training Accuracy : 0.8626702222423179
Test Accuracy : 0.7954868193972229
# Initialization to store accuracy value
train_accu = []
test_accu = []
- Linear Regression after Transform
lr.fit(x_train_transform1, y_train)
y_pred_train_ = lr.predict(x_train_transform1)
y_pred_test_ = lr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_))
print()
print("Test Accuracy : ",r2_score(y_test, y_pred_test_))
train_accu.append(r2_score(y_train,y_pred_train_))
test_accu.append(r2_score(y_test,y_pred_test_))
Training Accuracy : 0.8660561096838644
Test Accuracy : 0.8089635049182979
- Variance Inflation Factor (VIF) of Transformed Data
x1 = df_trans.copy()
from statsmodels.stats.outliers_influence import variance_inflation_factor
variable = x1
vif = pd.DataFrame()
vif['Variance Inflation Factor'] = [variance_inflation_factor(variable, i)
for i in range(variable.shape[1])]
vif['Features'] = x1.columns
x2 = x1.copy()
x2.drop(['Area'], axis = 1, inplace=True)
variable = x2
vif = pd.DataFrame()
vif['Variance Inflation Factor'] = [variance_inflation_factor(variable, i)
for i in range(variable.shape[1])]
vif['Features'] = x2.columns
print (vif)
Variance Inflation Factor Features
0 19.613495 Production
1 6.502925 Annual_Rainfall
2 18.511399 Fertilizer
3 4.398463 Crop_Arhar/Tur
4 4.615635 Crop_Bajra
.. ... ...
86 1.327546 State_Telangana
87 1.899844 State_Tripura
88 1.737332 State_Uttar Pradesh
89 1.901585 State_Uttarakhand
90 2.399254 State_West Bengal
[91 rows x 2 columns]
x2.drop(['Production'], axis = 1, inplace=True)
variable = x2
vif = pd.DataFrame()
vif['Variance Inflation Factor'] = [variance_inflation_factor(variable, i)
for i in range(variable.shape[1])]
vif['Features'] = x2.columns
print (vif)
Variance Inflation Factor Features
0 6.502762 Annual_Rainfall
1 2.228737 Fertilizer
2 4.391123 Crop_Arhar/Tur
3 4.615516 Crop_Bajra
4 2.518605 Crop_Banana
.. ... ...
85 1.325111 State_Telangana
86 1.895037 State_Tripura
87 1.737068 State_Uttar Pradesh
88 1.899269 State_Uttarakhand
89 2.387405 State_West Bengal
[90 rows x 2 columns]
- Linear Regression after Applying VIF
x_test1 = pd.DataFrame(x_test_transform1, columns=x_test.columns)
x_test1.drop(['Area','Production'], axis = 1, inplace = True)
# After applying vif
lr.fit(x2, y_train)
y_pred_train_ = lr.predict(x2)
y_pred_test_ = lr.predict(x_test1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_))
print()
print("Test Accuracy : ",r2_score(y_test, y_pred_test_))
train_accu.append(r2_score(y_train,y_pred_train_))
test_accu.append(r2_score(y_test,y_pred_test_))
Training Accuracy : 0.8612782567666339
Test Accuracy : 0.7993727827493086
- Random Forest Regressor of Transformed Data
from sklearn.ensemble import RandomForestRegressor
regr = RandomForestRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9976015686329384
Test Accuracy : 0.9453495882397677
- Random Forest Yield Test Predicted/Observed Transformed Data
plt.scatter(y_test,y_pred_test_regr)
plt.title(" RandomForest Yield Test Data")
plt.xlabel("Observed Data")
plt.ylabel("Predicted Data")
plt.grid()
plt.show()
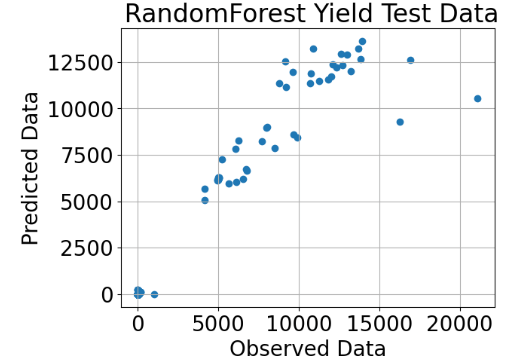
- Random Forest Regressor of Transformed Data after applying VIF
from sklearn.ensemble import RandomForestRegressor
regr = RandomForestRegressor()
regr.fit(x2, y_train)
y_pred_train_regr= regr.predict(x2)
y_pred_test_regr = regr.predict(x_test1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9968700195835773
Test Accuracy : 0.9512089658064634
- Random Forest Yield Test Predicted/Observed Transformed Data after applying VIF
plt.scatter(y_test,y_pred_test_regr)
plt.title(" RandomForest Yield Test Data")
plt.xlabel("Observed Data")
plt.ylabel("Predicted Data")
plt.grid()
plt.show()

- Let’s check the accuracy of various regression models
Linear Regression
from sklearn.linear_model import LinearRegression
regr = LinearRegression()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.8660561096838644
Test Accuracy : 0.8089635049182979
Ada Boost Regressor
from sklearn.ensemble import AdaBoostRegressor
regr = AdaBoostRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9753826862500993
Test Accuracy : 0.9432430977757433
Decision Tree Regressor
from sklearn.tree import DecisionTreeRegressor
regr = DecisionTreeRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 1.0
Test Accuracy : 0.953298449231916
Extra Trees Regressor
from sklearn.ensemble import ExtraTreesRegressor
regr = ExtraTreesRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 1.0
Test Accuracy : 0.9252164492737693
Gradient Boosting Regressor
from sklearn.ensemble import GradientBoostingRegressor
regr = GradientBoostingRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9981826164721128
Test Accuracy : 0.9057446023833922
MLP Regressor
from sklearn.neural_network import MLPRegressor
regr = MLPRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9590513944201091
Test Accuracy : 0.8870588171504107
K-Neighbors Regressor
from sklearn.neighbors import KNeighborsRegressor
regr = KNeighborsRegressor()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.9851698885429
Test Accuracy : 0.9387721214512218
Elastic Net CV
from sklearn.linear_model import ElasticNetCV
regr = ElasticNetCV()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.7017372197891507
Test Accuracy : 0.6619959299722024
Kernel Ridge
from sklearn.kernel_ridge import KernelRidge
regr = KernelRidge()
regr.fit(x_train_transform1, y_train)
y_pred_train_regr= regr.predict(x_train_transform1)
y_pred_test_regr = regr.predict(x_test_transform1)
print("Training Accuracy : ",r2_score(y_train, y_pred_train_regr))
print("Test Accuracy : ",r2_score(y_test, y_pred_test_regr))
train_accu.append(r2_score(y_train,y_pred_train_regr))
test_accu.append(r2_score(y_test,y_pred_test_regr))
Training Accuracy : 0.8574072843636276
Test Accuracy : 0.8015219127341822
| Train/Test Data | Algorithm | R2-Score Train | R2-Score Test |
| Before Transform | Linear Regression | 0.79 | |
| After Transform | Linear Regression | 0.86 | 0.81 |
| After Transform + VIF | Linear Regression | 0.86 | 0.80 |
| After Transform | Random Forest Regressor | 0.99 | 0.94 |
| After Transform + VIF | Random Forest Regressor | 0.99 | 0.95 |
| After Transform | Ada Boost Regressor | 0.97 | 0.94 |
| After Transform | Decision Tree Regressor | 1.00 | 0.95 |
| After Transform | Gradient Boosting Regressor | 0.99 | 0.90 |
| After Transform | MLP Regressor | 0.96 | 0.89 |
| After Transform | KNeighbors Regressor | 0.98 | 0.94 |
| After Transform | Extra Trees Regressor | 1.00 | 0.92 |
| After Transform | Elastic Net CV | 0.70 | 0.66 |
| After Transform | Kernel Ridge | 0.86 | 0.80 |
- Random Forest Regressor yields the best model in terms of the R2-score. Both Decision Tree and Extra Trees Regressors suffer from training data overfitting. The Elastic Net CV provides the worst model closest to a random guess (null model).
Global Crop Yield Prediction
- Agriculture plays a critical role in the global economy. With the continuing expansion of the human population understanding worldwide crop yield is central to addressing food security challenges and reducing the impacts of climate change.
- In this section, we will focus on top 10 most consumed yields all over the world to predict top yields globally by applying supervised ML techniques.
- These crops of interest are Cassava, Maize, Plantains and others, Potatoes, Rice, Paddy, Sorghum, Soybeans, Sweet Potatoes, Wheat, and Yams
- The input dataset DAT2 consists of the 4 CSV files: pesticides, rainfall, temp, and yield. Here, Pesticides & Yield are collected from FAO; Rainfall & Avg. Temperature are collected from World Data Bank.
- Input data loading, editing, and preliminary analysis
import numpy as np
import pandas as pd
df_yield = pd.read_csv('yield.csv')
# rename columns.
df_yield = df_yield.rename(index=str, columns={"Value": "hg/ha_yield"})
# drop unwanted columns.
df_yield = df_yield.drop(['Year Code','Element Code','Element','Year Code','Area Code','Domain Code','Domain','Unit','Item Code'], axis=1)
df_rain = pd.read_csv('rainfall.csv')
df_rain = df_rain.rename(index=str, columns={" Area": 'Area'})
df_rain['average_rain_fall_mm_per_year'] = pd.to_numeric(df_rain['average_rain_fall_mm_per_year'],errors = 'coerce')
df_rain = df_rain.dropna()
yield_df = pd.merge(df_yield, df_rain, on=['Year','Area'])
df_pes = pd.read_csv('pesticides.csv')
f_pes = df_pes.rename(index=str, columns={"Value": "pesticides_tonnes"})
df_pes = df_pes.drop(['Element','Domain','Unit','Item'], axis=1)
yield_df = pd.merge(yield_df, df_pes, on=['Year','Area'])
avg_temp= pd.read_csv('temp.csv')
avg_temp = avg_temp.rename(index=str, columns={"year": "Year", "country":'Area'})
yield_df = pd.merge(yield_df,avg_temp, on=['Area','Year'])
yield_df.groupby('Item').count()

yield_df.groupby(['Area'],sort=True)['hg/ha_yield'].sum().nlargest(10)
Area
India 327420324
Brazil 167550306
Mexico 130788528
Japan 124470912
Australia 109111062
Pakistan 73897434
Indonesia 69193506
United Kingdom 55419990
Turkey 52263950
Spain 46773540
Name: hg/ha_yield, dtype: int64
yield_df.groupby(['Item','Area'],sort=True)['hg/ha_yield'].sum().nlargest(10)
Item Area
Cassava India 142810624
Potatoes India 92122514
Brazil 49602168
United Kingdom 46705145
Australia 45670386
Sweet potatoes India 44439538
Potatoes Japan 42918726
Mexico 42053880
Sweet potatoes Mexico 35808592
Australia 35550294
Name: hg/ha_yield, dtype: int64
- Feature correlation analysis
import sklearn
import seaborn as sns
import matplotlib.pyplot as plt
correlation_data=yield_df.select_dtypes(include=[np.number]).corr()
mask = np.zeros_like(correlation_data, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
cmap = sns.palette="vlag"
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(correlation_data, mask=mask, cmap=cmap, vmax=.3, center=0,annot=True,
square=True, linewidths=.5, cbar_kws={"shrink": .5});
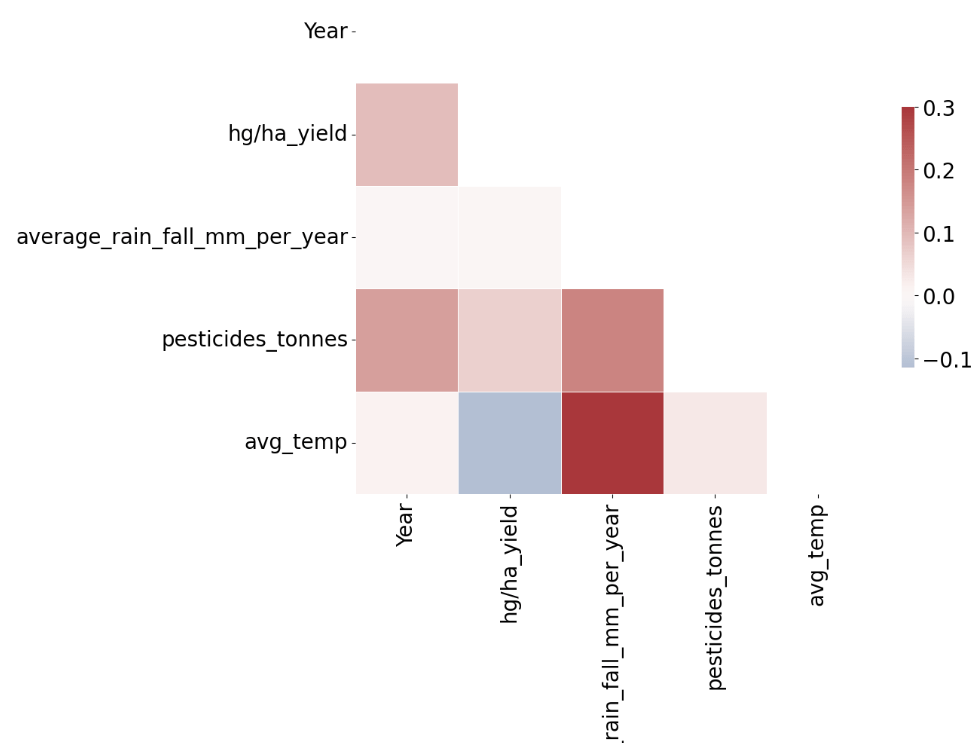
- Data preparation for ML – One Hot Encoder, Min/Max Scaling, and Train/Test Split
from sklearn.preprocessing import OneHotEncoder
yield_df_onehot = pd.get_dummies(yield_df, columns=['Area',"Item"], prefix = ['Country',"Item"])
features=yield_df_onehot.loc[:, yield_df_onehot.columns != 'hg/ha_yield']
label=yield_df['hg/ha_yield']
features = features.drop(['Year'], axis=1)
from sklearn.preprocessing import MinMaxScaler
scaler=MinMaxScaler()
features=scaler.fit_transform(features)
from sklearn.model_selection import train_test_split
train_data, test_data, train_labels, test_labels = train_test_split(features, label, test_size=0.2, random_state=42)
- ML model training and testing
from sklearn.metrics import r2_score
def compare_models(model):
model_name = model.__class__.__name__
fit=model.fit(train_data,train_labels)
y_pred=fit.predict(test_data)
r2=r2_score(test_labels,y_pred)
return([model_name,r2])
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn import svm
from sklearn.tree import DecisionTreeRegressor
models = [
GradientBoostingRegressor(n_estimators=200, max_depth=3, random_state=0),
RandomForestRegressor(n_estimators=200, max_depth=3, random_state=0),
svm.SVR(),
DecisionTreeRegressor()
]
model_train=list(map(compare_models,models))
print(*model_train, sep = "\n")
['GradientBoostingRegressor', 0.8959545600619471]
['RandomForestRegressor', 0.6807690552605921]
['SVR', -0.19466686625412555]
['DecisionTreeRegressor', 0.959733560005363]
We select Decision Tree Regressor for ML training and testing.
yield_df_onehot = yield_df_onehot.drop(['Year'], axis=1)
#setting test data to columns from dataframe and excluding 'hg/ha_yield' values where ML model should be predicting
test_df=pd.DataFrame(test_data,columns=yield_df_onehot.loc[:, yield_df_onehot.columns != 'hg/ha_yield'].columns)
# using stack function to return a reshaped DataFrame by pivoting the columns of the current dataframe
cntry=test_df[[col for col in test_df.columns if 'Country' in col]].stack()[test_df[[col for col in test_df.columns if 'Country' in col]].stack()>0]
cntrylist=list(pd.DataFrame(cntry).index.get_level_values(1))
countries=[i.split("_")[1] for i in cntrylist]
itm=test_df[[col for col in test_df.columns if 'Item' in col]].stack()[test_df[[col for col in test_df.columns if 'Item' in col]].stack()>0]
itmlist=list(pd.DataFrame(itm).index.get_level_values(1))
items=[i.split("_")[1] for i in itmlist]
test_df.drop([col for col in test_df.columns if 'Item' in col],axis=1,inplace=True)
test_df.drop([col for col in test_df.columns if 'Country' in col],axis=1,inplace=True)
test_df['Country']=countries
test_df['Item']=items
from sklearn.tree import DecisionTreeRegressor
clf=DecisionTreeRegressor()
model=clf.fit(train_data,train_labels)
test_df["yield_predicted"]= model.predict(test_data)
test_df["yield_actual"]=pd.DataFrame(test_labels)["hg/ha_yield"].tolist()
test_group=test_df.groupby("Item")
# So let's run the model actual values against the predicted ones
fig, ax = plt.subplots()
ax.scatter(test_df["yield_actual"], test_df["yield_predicted"],edgecolors=(0, 0, 0))
ax.set_xlabel('Actual')
ax.set_ylabel('Predicted')
ax.set_title("DecisionTree Yield Test Data")
x=test_df["yield_actual"]
y=test_df["yield_predicted"]
m, b = np.polyfit(x, y, 1)
#add linear regression line to scatterplot
plt.plot(x, m*x+b,'r',lw=2)
plt.grid()
plt.show()
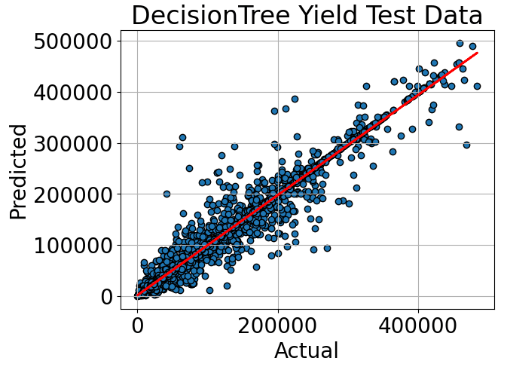
- Feature Importance Factor
varimp= {'imp':model.feature_importances_,'names':yield_df_onehot.columns[yield_df_onehot.columns!="hg/ha_yield"]}
a4_dims = (8.27,19)
plt.rcParams.update({'font.size': 10})
fig, ax = plt.subplots(figsize=a4_dims)
df=pd.DataFrame.from_dict(varimp)
df.sort_values(ascending=False,by=["imp"],inplace=True)
plt.title('Imp')
df=df.dropna()
sns.barplot(x="imp",y="names",palette="vlag",data=df,orient="h",ax=ax);
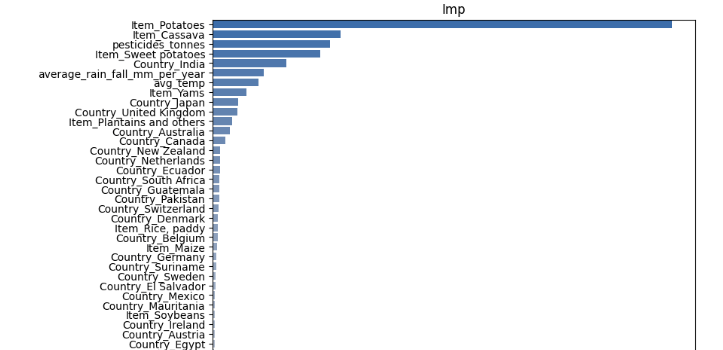
0.00 0.35
#7 most important factors that affect crops
a4_dims = (16.7, 8.27)
plt.rcParams.update({'font.size': 20})
fig, ax = plt.subplots(figsize=a4_dims)
df=pd.DataFrame.from_dict(varimp)
df.sort_values(ascending=False,by=["imp"],inplace=True)
df=df.dropna()
df=df.nlargest(7, 'imp')
sns.barplot(x="imp",y="names",palette="vlag",data=df,orient="h",ax=ax);
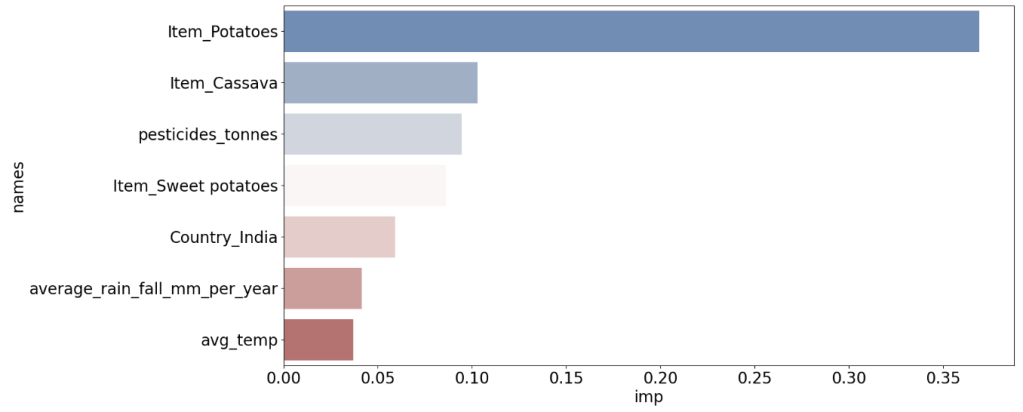
- Global Yield Prediction
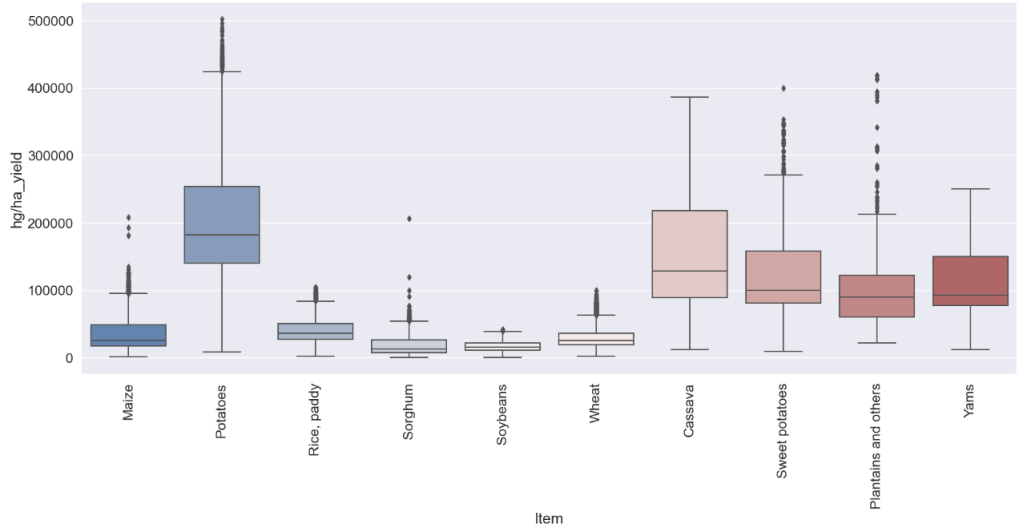
#Boxplot that shows yield for each item
a4_dims = (20, 8)
fig, ax = plt.subplots(figsize=a4_dims)
sns.boxplot(x="Item",y="hg/ha_yield",palette="vlag",data=yield_df,ax=ax);
plt.xticks(rotation=90)
plt.savefig('yield_boxplot.png')
Crop Recommendation
- Here, we will implement the multi-label crop classification algorithm using the crop recommendation dataset DAT3. This is an important decision support tool for crop yield prediction, including supporting decisions on what crops to grow and what to do during the growing season of the crops.
- Key imports and input data loading
import pandas as pd # data analysis
import numpy as np # linear algebra
#import libraries for data visualization
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
import plotly.graph_objects as go
import plotly.express as px
from plotly.subplots import make_subplots
from sklearn.metrics import classification_report
from sklearn import metrics
from sklearn import tree
from sklearn.model_selection import cross_val_score
import warnings
warnings.filterwarnings('ignore')
crop = pd.read_csv('Crop_recommendation.csv')
- Exploratory Data Analysis (EDA)
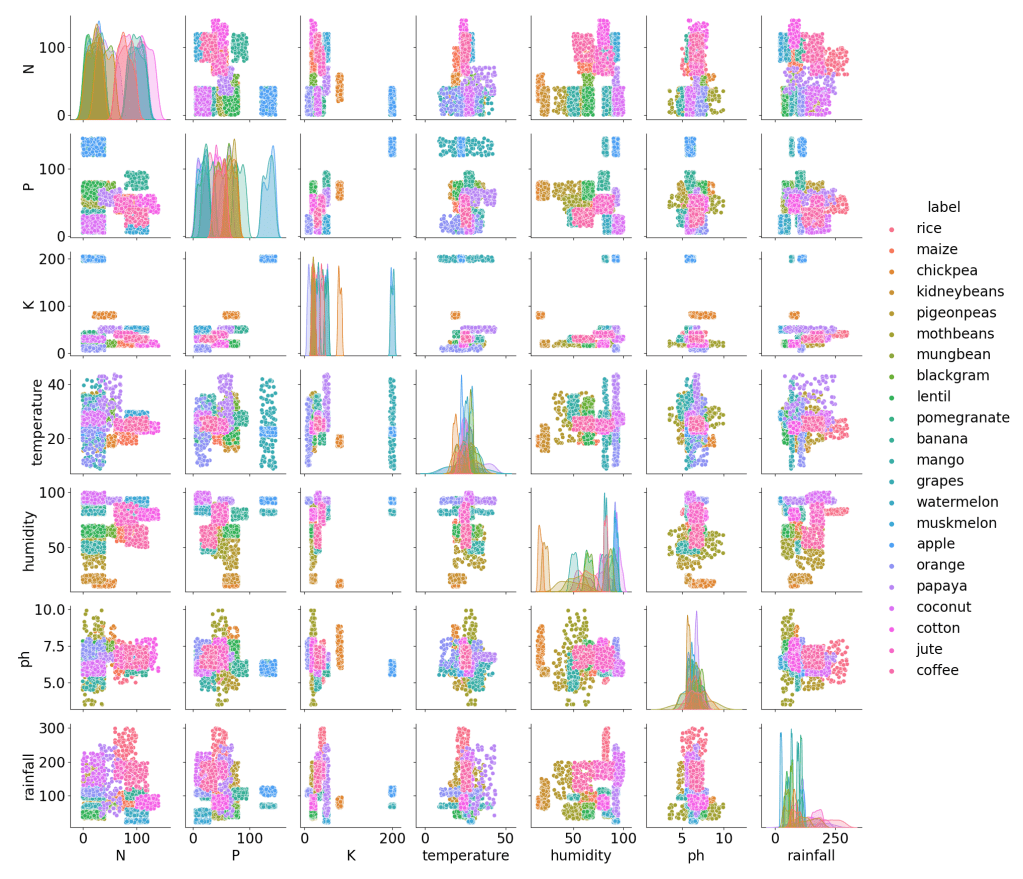
Pair plot of available features (K,P,N, temperature, humidity, pH, and rainfall) for different crop labels:
sns.pairplot(crop,hue = 'label')
plt.savefig('crop_pairplot.png')
Joint plot humidity vs rainfall for different crop labels:
plt.figure(figsize=(10,10))
sns.jointplot(x="rainfall",y="humidity",data=crop[(crop['temperature']<40) &
(crop['rainfall']>40)],height=10,hue="label")
plt.rcParams.update({'font.size': 12})
plt.savefig('crop_jointplot.png')

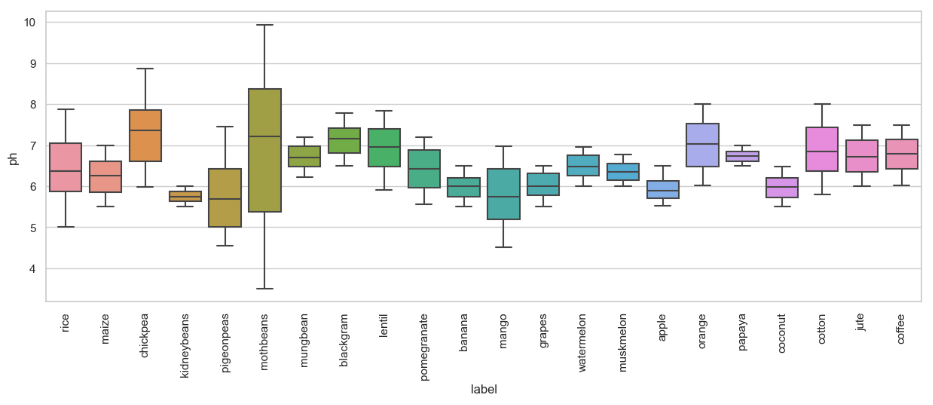
Box plots pH level vs crop labels:
sns.set_theme(style="whitegrid")
fig, ax = plt.subplots(figsize=(15,5))
sns.set(font_scale=1.5)
sns.boxplot(x='label',y='ph',data=crop)
plt.rcParams.update({'font.size': 22})
plt.xticks(rotation=90)
(array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21]),
[Text(0, 0, 'rice'),
Text(1, 0, 'maize'),
Text(2, 0, 'chickpea'),
Text(3, 0, 'kidneybeans'),
Text(4, 0, 'pigeonpeas'),
Text(5, 0, 'mothbeans'),
Text(6, 0, 'mungbean'),
Text(7, 0, 'blackgram'),
Text(8, 0, 'lentil'),
Text(9, 0, 'pomegranate'),
Text(10, 0, 'banana'),
Text(11, 0, 'mango'),
Text(12, 0, 'grapes'),
Text(13, 0, 'watermelon'),
Text(14, 0, 'muskmelon'),
Text(15, 0, 'apple'),
Text(16, 0, 'orange'),
Text(17, 0, 'papaya'),
Text(18, 0, 'coconut'),
Text(19, 0, 'cotton'),
Text(20, 0, 'jute'),
Text(21, 0, 'coffee')])
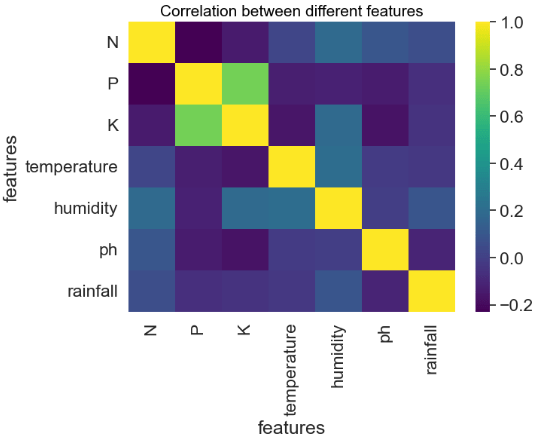
- Feature Correlation Matrix
Correlation between different features:
fig, ax = plt.subplots(1, 1, figsize=(7, 5))
plt.rcParams.update({'font.size': 20})
sns.set(font_scale=1.5)
sns.heatmap(crop.corr(), annot=False,cmap='viridis')
ax.set(xlabel='features')
ax.set(ylabel='features')
plt.title('Correlation between different features', fontsize = 15, c='black')
plt.savefig('corrmatrix0.png')
- Crop Summary Bar Plot
N-P-K values comparison between crops:
crop_summary = pd.pivot_table(crop,index=['label'],aggfunc='mean')
fig = go.Figure()
fig.add_trace(go.Bar(
x=crop_summary.index,
y=crop_summary['N'],
name='Nitrogen',
marker_color='mediumvioletred'
))
fig.add_trace(go.Bar(
x=crop_summary.index,
y=crop_summary['P'],
name='Phosphorous',
marker_color='springgreen'
))
fig.add_trace(go.Bar(
x=crop_summary.index,
y=crop_summary['K'],
name='Potash',
marker_color='dodgerblue'
))
fig.update_layout(title="N-P-K values comparison between crops",
plot_bgcolor='white',
barmode='group',
xaxis_tickangle=-45)
fig.show()

- ML Data Preparation
features = crop[['N', 'P','K','temperature', 'humidity', 'ph', 'rainfall']]
target = crop['label']
acc = []
model = []
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(features,target,test_size = 0.3,random_state =2)
- K-Neighbors (KNN) Classifier
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier()
knn.fit(x_train,y_train)
predicted_values = knn.predict(x_test)
x = metrics.accuracy_score(y_test, predicted_values)
acc.append(x)
model.append('K Nearest Neighbours')
print("KNN Accuracy is: ", x)
print(classification_report(y_test,predicted_values))
KNN Accuracy is: 0.9848484848484849
precision recall f1-score support
apple 1.00 1.00 1.00 28
banana 1.00 1.00 1.00 26
blackgram 1.00 1.00 1.00 28
chickpea 1.00 1.00 1.00 29
coconut 1.00 1.00 1.00 31
coffee 1.00 1.00 1.00 33
cotton 0.97 1.00 0.98 31
grapes 1.00 1.00 1.00 29
jute 0.91 0.88 0.89 33
kidneybeans 0.97 1.00 0.98 30
lentil 0.97 1.00 0.98 32
maize 1.00 0.97 0.98 32
mango 1.00 1.00 1.00 33
mothbeans 1.00 0.97 0.98 29
mungbean 1.00 1.00 1.00 32
muskmelon 1.00 1.00 1.00 30
orange 1.00 1.00 1.00 42
papaya 1.00 1.00 1.00 30
pigeonpeas 1.00 0.97 0.98 31
pomegranate 1.00 1.00 1.00 19
rice 0.87 0.90 0.89 30
watermelon 1.00 1.00 1.00 22
accuracy 0.98 660
macro avg 0.99 0.99 0.99 660
weighted avg 0.99 0.98 0.98 660
score = cross_val_score(knn,features,target,cv=5)
print('Cross validation score: ',score)
Cross validation score: [0.97727273 0.98181818 0.97954545 0.97954545 0.97954545]
#Print Train Accuracy
knn_train_accuracy = knn.score(x_train,y_train)
print("knn_train_accuracy = ",knn.score(x_train,y_train))
#Print Test Accuracy
knn_test_accuracy = knn.score(x_test,y_test)
print("knn_test_accuracy = ",knn.score(x_test,y_test))
knn_train_accuracy = 0.9876623376623377
knn_test_accuracy = 0.9848484848484849
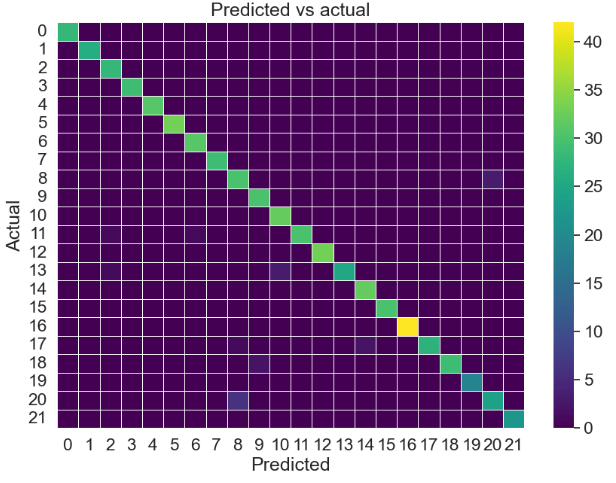
KNN confusion matrix:
y_pred = knn.predict(x_test)
y_true = y_test
from sklearn.metrics import confusion_matrix
cm_knn = confusion_matrix(y_true,y_pred)
f, ax = plt.subplots(figsize=(10,7))
sns.set(font_scale=1.5)
sns.heatmap(cm_knn, annot=False, linewidth=0.5, fmt=".0f",cmap='viridis', ax = ax)
plt.xlabel('Predicted')
plt.ylabel('Actual')
plt.title('Predicted vs actual')
plt.show()
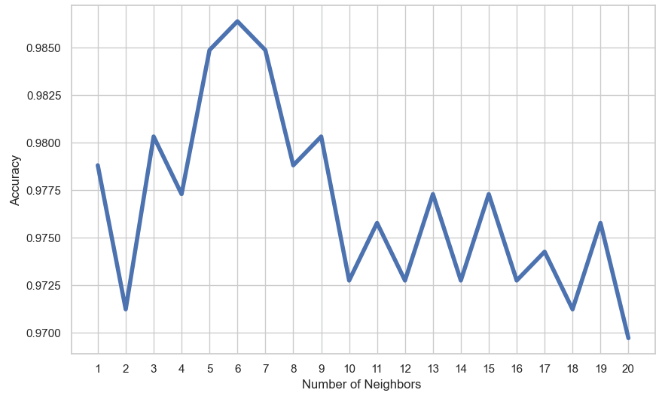
KNN Accuracy vs Number of Neighbors:
mean_acc = np.zeros(20)
for i in range(1,21):
#Train Model and Predict
knn = KNeighborsClassifier(n_neighbors = i).fit(x_train,y_train)
yhat= knn.predict(x_test)
mean_acc[i-1] = metrics.accuracy_score(y_test, yhat)
loc = np.arange(1,21,step=1.0)
plt.figure(figsize = (10, 6))
plt.plot(range(1,21), mean_acc,lw=4)
plt.xticks(loc)
plt.xlabel('Number of Neighbors ')
plt.ylabel('Accuracy')
plt.show()
- KNN HPO via Grid Search CV
from sklearn.model_selection import GridSearchCV
grid_params = { 'n_neighbors' : [6,9,11,13,15,17,19],
'weights' : ['uniform','distance'],
'metric' : ['minkowski','euclidean','manhattan']}
gs = GridSearchCV(KNeighborsClassifier(), grid_params, verbose = 1, cv=3, n_jobs = -1)
g_res = gs.fit(x_train, y_train)
Fitting 3 folds for each of 42 candidates, totalling 126 fits
g_res.best_score_
0.9844143071325814
g_res.best_params_
{'metric': 'manhattan', 'n_neighbors': 6, 'weights': 'distance'}
# Using the best hyperparameters
knn_1 = KNeighborsClassifier(n_neighbors = 6, weights = 'distance',algorithm = 'brute',metric = 'manhattan')
knn_1.fit(x_train, y_train)
KNeighborsClassifier
KNeighborsClassifier(algorithm='brute', metric='manhattan', n_neighbors=6,
weights='distance')
# Training & Testing accuracy after applying hyper parameter
knn_train_accuracy = knn_1.score(x_train,y_train)
print("knn_train_accuracy = ",knn_1.score(x_train,y_train))
#Print Test Accuracy
knn_test_accuracy = knn_1.score(x_test,y_test)
print("knn_test_accuracy = ",knn_1.score(x_test,y_test))
knn_train_accuracy = 1.0
knn_test_accuracy = 0.9818181818181818
- Decision Tree (DT) Classifier
from sklearn.tree import DecisionTreeClassifier
DT = DecisionTreeClassifier(criterion="entropy",random_state=2,max_depth=5)
DT.fit(x_train,y_train)
predicted_values = DT.predict(x_test)
x = metrics.accuracy_score(y_test, predicted_values)
acc.append(x)
model.append('Decision Tree')
print("Decision Tree's Accuracy is: ", x*100)
print(classification_report(y_test,predicted_values))
Decision Tree's Accuracy is: 85.9090909090909
precision recall f1-score support
apple 1.00 1.00 1.00 28
banana 1.00 1.00 1.00 26
blackgram 0.65 1.00 0.79 28
chickpea 1.00 1.00 1.00 29
coconut 0.94 1.00 0.97 31
coffee 1.00 1.00 1.00 33
cotton 1.00 0.97 0.98 31
grapes 1.00 1.00 1.00 29
jute 0.50 0.03 0.06 33
kidneybeans 0.00 0.00 0.00 30
lentil 0.63 1.00 0.77 32
maize 0.97 1.00 0.98 32
mango 1.00 1.00 1.00 33
mothbeans 0.00 0.00 0.00 29
mungbean 1.00 1.00 1.00 32
muskmelon 1.00 1.00 1.00 30
orange 1.00 1.00 1.00 42
papaya 1.00 1.00 1.00 30
pigeonpeas 0.55 1.00 0.71 31
pomegranate 1.00 1.00 1.00 19
rice 0.49 0.97 0.65 30
watermelon 1.00 1.00 1.00 22
accuracy 0.86 660
macro avg 0.81 0.86 0.81 660
weighted avg 0.80 0.86 0.81 660
score = cross_val_score(DT, features, target,cv=5)
print('Cross validation score: ',score)
Cross validation score: [0.93636364 0.90909091 0.91818182 0.87045455 0.93636364]
#Print Train Accuracy
dt_train_accuracy = DT.score(x_train,y_train)
print("Training accuracy = ",DT.score(x_train,y_train))
#Print Test Accuracy
dt_test_accuracy = DT.score(x_test,y_test)
print("Testing accuracy = ",DT.score(x_test,y_test))
Training accuracy = 0.8675324675324675
Testing accuracy = 0.8590909090909091
Decision Tree confusion matrix:
y_pred = DT.predict(x_test)
y_true = y_test
from sklearn.metrics import confusion_matrix
cm_dt = confusion_matrix(y_true,y_pred)
f, ax = plt.subplots(figsize=(10,7))
sns.set(font_scale=1.5)
sns.heatmap(cm_dt, annot=False, linewidth=0.5, fmt=".0f", cmap='viridis', ax = ax)
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title('Predicted vs actual')
plt.show()

- Random Forest (RF) Classifier
from sklearn.ensemble import RandomForestClassifier
RF = RandomForestClassifier(n_estimators=20, random_state=0)
RF.fit(x_train,y_train)
predicted_values = RF.predict(x_test)
x = metrics.accuracy_score(y_test, predicted_values)
acc.append(x)
model.append('RF')
print("Random Forest Accuracy is: ", x)
print(classification_report(y_test,predicted_values))
Random Forest Accuracy is: 0.9893939393939394
precision recall f1-score support
apple 1.00 1.00 1.00 28
banana 1.00 1.00 1.00 26
blackgram 0.97 1.00 0.98 28
chickpea 1.00 1.00 1.00 29
coconut 1.00 1.00 1.00 31
coffee 1.00 1.00 1.00 33
cotton 1.00 1.00 1.00 31
grapes 1.00 1.00 1.00 29
jute 0.89 0.94 0.91 33
kidneybeans 1.00 1.00 1.00 30
lentil 1.00 1.00 1.00 32
maize 1.00 1.00 1.00 32
mango 1.00 1.00 1.00 33
mothbeans 1.00 0.97 0.98 29
mungbean 1.00 1.00 1.00 32
muskmelon 1.00 1.00 1.00 30
orange 1.00 1.00 1.00 42
papaya 1.00 1.00 1.00 30
pigeonpeas 1.00 1.00 1.00 31
pomegranate 1.00 1.00 1.00 19
rice 0.93 0.87 0.90 30
watermelon 1.00 1.00 1.00 22
accuracy 0.99 660
macro avg 0.99 0.99 0.99 660
weighted avg 0.99 0.99 0.99 660
score = cross_val_score(RF,features,target,cv=5)
print('Cross validation score: ',score)
Cross validation score: [0.99772727 0.99545455 0.99772727 0.99318182 0.98863636]
#Print Train Accuracy
rf_train_accuracy = RF.score(x_train,y_train)
print("Training accuracy = ",RF.score(x_train,y_train))
#Print Test Accuracy
rf_test_accuracy = RF.score(x_test,y_test)
print("Testing accuracy = ",RF.score(x_test,y_test))
Training accuracy = 1.0
Testing accuracy = 0.9893939393939394
Random Forest confusion matrix:
y_pred = RF.predict(x_test)
y_true = y_test
from sklearn.metrics import confusion_matrix
cm_rf = confusion_matrix(y_true,y_pred)
f, ax = plt.subplots(figsize=(10,7))
sns.set(font_scale=1.5)
sns.heatmap(cm_rf, annot=False, linewidth=0.5, fmt=".0f", cmap='viridis', ax = ax)
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title('Predicted vs actual')
plt.show()

- Gaussian NB Classifier
from sklearn.naive_bayes import GaussianNB
NaiveBayes = GaussianNB()
NaiveBayes.fit(x_train,y_train)
predicted_values = NaiveBayes.predict(x_test)
x = metrics.accuracy_score(y_test, predicted_values)
acc.append(x)
model.append('Naive Bayes')
print("Naive Bayes Accuracy is: ", x)
print(classification_report(y_test,predicted_values))
Naive Bayes Accuracy is: 0.9924242424242424
precision recall f1-score support
apple 1.00 1.00 1.00 28
banana 1.00 1.00 1.00 26
blackgram 1.00 1.00 1.00 28
chickpea 1.00 1.00 1.00 29
coconut 1.00 1.00 1.00 31
coffee 1.00 1.00 1.00 33
cotton 1.00 1.00 1.00 31
grapes 1.00 1.00 1.00 29
jute 0.89 0.97 0.93 33
kidneybeans 1.00 1.00 1.00 30
lentil 1.00 1.00 1.00 32
maize 1.00 1.00 1.00 32
mango 1.00 1.00 1.00 33
mothbeans 1.00 1.00 1.00 29
mungbean 1.00 1.00 1.00 32
muskmelon 1.00 1.00 1.00 30
orange 1.00 1.00 1.00 42
papaya 1.00 1.00 1.00 30
pigeonpeas 1.00 1.00 1.00 31
pomegranate 1.00 1.00 1.00 19
rice 0.96 0.87 0.91 30
watermelon 1.00 1.00 1.00 22
accuracy 0.99 660
macro avg 0.99 0.99 0.99 660
weighted avg 0.99 0.99 0.99 660
score = cross_val_score(NaiveBayes,features,target,cv=5)
print('Cross validation score: ',score)
Cross validation score: [0.99772727 0.99545455 0.99545455 0.99545455 0.99090909]
#Print Train Accuracy
nb_train_accuracy = NaiveBayes.score(x_train,y_train)
print("Training accuracy = ",NaiveBayes.score(x_train,y_train))
#Print Test Accuracy
nb_test_accuracy = NaiveBayes.score(x_test,y_test)
print("Testing accuracy = ",NaiveBayes.score(x_test,y_test))
Training accuracy = 0.9961038961038962
Testing accuracy = 0.9924242424242424
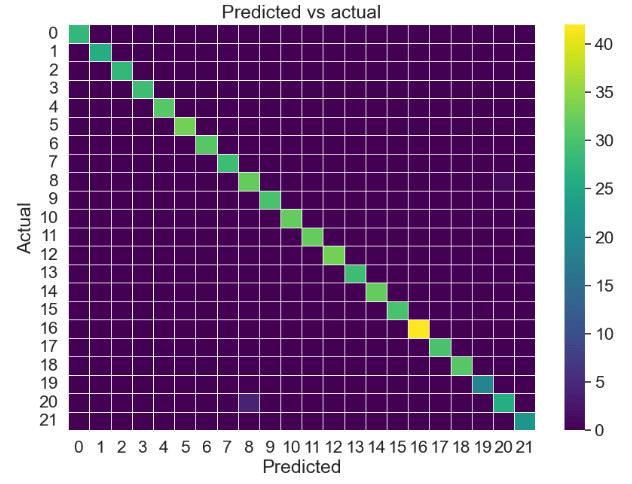
Naïve Bayes confusion matrix:
y_pred = NaiveBayes.predict(x_test)
y_true = y_test
sns.set(font_scale=1.5)
from sklearn.metrics import confusion_matrix
cm_nb = confusion_matrix(y_true,y_pred)
f, ax = plt.subplots(figsize=(10,7))
sns.heatmap(cm_nb, annot=False, linewidth=0.5, fmt=".0f", cmap='viridis', ax = ax)
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title('Predicted vs actual')
plt.show()
- Test ML Accuracy Score Bar Plot
plt.figure(figsize=[14,7],dpi = 100, facecolor='white')
sns.set(font_scale=2)
plt.title('Accuracy Comparison')
plt.xlabel('Accuracy')
plt.ylabel('ML Algorithms')
sns.barplot(x = acc,y = model,palette='viridis')
plt.savefig('plot.png', dpi=300, bbox_inches='tight')

- Testing vs Training ML Accuracy Comparison Bar Plot
label = ['KNN', 'Decision Tree','Random Forest','Naive Bayes']
Test = [knn_test_accuracy, dt_test_accuracy,rf_test_accuracy,
nb_test_accuracy]
Train = [knn_train_accuracy, dt_train_accuracy, rf_train_accuracy,
nb_train_accuracy]
f, ax = plt.subplots(figsize=(20,7)) # set the size that you'd like (width, height)
X_axis = np.arange(len(label))
plt.bar(X_axis - 0.2,Test, 0.4, label = 'Test', color=('midnightblue'))
plt.bar(X_axis + 0.2,Train, 0.4, label = 'Train', color=('mediumaquamarine'))
plt.rcParams.update({'font.size': 20})
plt.xticks(X_axis, label)
plt.xlabel("ML algorithms")
plt.ylabel("Accuracy")
plt.title("Testing vs Training Accuracy")
plt.legend()
#plt.savefig('train vs test.png')
plt.show()

Effects of Climate Changes
- Finally, we will examine the effect of climate changes on crop yields worldwide.
- The input dataset DAT4 describes Effects of Climate Changes on Crop Yields worldwide.
- This dataset has 7 columns and 28242 rows.
Area
Item
Year
average_rain_fall_mm_per_year
pesticides_tonnes
avg_temp
hg/ha_yield
- Basic imports and load the input dataset
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
import seaborn as sns
%matplotlib inline
df=pd.read_csv('climate-ds.csv',index_col=0)
- Evaluation functions for regression models
from sklearn.metrics import mean_absolute_error, mean_squared_error, max_error, median_absolute_error, r2_score, explained_variance_score, mean_absolute_percentage_error
import numpy as np
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
#Evaluation function for regression models
def regression_report(y_true, y_pred):
error = y_true - y_pred
#Evaluation matrics
mae = mean_absolute_error(y_true, y_pred)
medae = median_absolute_error(y_true, y_pred)
mse = mean_squared_error(y_true, y_pred)
rmse = np.sqrt(mean_squared_error(y_true, y_pred))
maxerr = max_error(y_true, y_pred)
r_squared = r2_score(y_true, y_pred)
evs = explained_variance_score(y_true, y_pred)
mape = mean_absolute_percentage_error(y_true, y_pred)
metrics = [
('Mean Absolute Error', mae),
('Median Absolute Error', medae),
('Mean Squared Error', mse),
('Root Mean Squared Error', rmse),
('Max error', maxerr) ,
('R2 score', r_squared),
('Explained variance score', evs),
('Mean Absolute Percentage Error', mape)
]
print('Regression Report:')
for metric_name, metric_value in metrics:
print(f'\t\t\t{metric_name:30s}: {metric_value: >20.3f}')
return mae, medae, mse, rmse, maxerr, r_squared, evs, mape
- ML data preparation
#remove Duplicates
df.drop_duplicates(keep='first',inplace = True)
#one hot encoding for the categorical columns to can be trained in the model
df = pd.get_dummies(df,columns=['Area','Item'])
df.rename(columns={x:x[5:] for x in df.columns[6:]}, inplace = True)
#split the data into the target and labels
x = df.drop(labels=['hg/ha_yield'], axis=1)
y = df['hg/ha_yield']
#Scaling
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
x = scaler.fit_transform(x)
#Split the data into the train and test datasets as 80:20 %
#split data to x_train, x_test, y_train, y_test
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.20, random_state=40,shuffle=True)
#Make DataFrame for track model accuracy
df_models = pd.DataFrame(columns=["Model", "MAE","MEDAE","MSE","RMSE", "Max Error","R2 Score","EVS","MAPE"])
- LGBM Regressor + Randomized Search CV
import lightgbm as lgb
import scipy as sp
lgbm = lgb.LGBMRegressor(
objective='regression',
n_jobs=1,
)
#dictionary for hyper parameters
param_dist = {'boosting_type': ['gbdt', 'dart', 'rf'],
'num_leaves': sp.stats.randint(2, 1001),
'subsample_for_bin': sp.stats.randint(10, 1001),
'min_split_gain': sp.stats.uniform(0, 5.0),
'min_child_weight': sp.stats.uniform(1e-6, 1e-2),
'reg_alpha': sp.stats.uniform(0, 1e-2),
'reg_lambda': sp.stats.uniform(0, 1e-2),
'tree_learner': ['data', 'feature', 'serial', 'voting' ],
'application': ['regression_l1', 'regression_l2', 'regression'],
'bagging_freq': sp.stats.randint(1, 11),
'bagging_fraction': sp.stats.uniform(1e-3, 0.99),
'feature_fraction': sp.stats.uniform(1e-3, 0.99),
'learning_rate': sp.stats.uniform(1e-6, 0.99),
'max_depth': sp.stats.randint(1, 501),
'n_estimators': sp.stats.randint(100, 20001),
}
rscv = RandomizedSearchCV(
estimator=lgbm,
param_distributions=param_dist,
cv=3,
verbose=0
)
rscv.fit(x_train, y_train)
print("The best parameters are %s with a score of %0.2f"
% (rscv.best_params_, rscv.best_score_))
gbr_pred = rscv.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, gbr_pred)
row = {"Model": "GBRegressor", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
The best parameters are {'application': 'regression_l2', 'bagging_fraction': 0.32615789951845936, 'bagging_freq': 5, 'boosting_type': 'gbdt', 'feature_fraction': 0.6168548991646009, 'learning_rate': 0.10299929691491512, 'max_depth': 442, 'min_child_weight': 0.007374638179549023, 'min_split_gain': 1.8189493243011623, 'n_estimators': 10896, 'num_leaves': 128, 'reg_alpha': 0.007666291745158882, 'reg_lambda': 0.0004935270085926446, 'subsample_for_bin': 63, 'tree_learner': 'voting'} with a score of 0.97
Regression Report:
Mean Absolute Error : 5611.731
Median Absolute Error : 2451.671
Mean Squared Error : 119179650.226
Root Mean Squared Error : 10916.943
Max error : 140538.210
R2 score : 0.984
Explained variance score : 0.984
Mean Absolute Percentage Error: 0.166
Gradient Boosting Regressor: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(12,5))
plt.rcParams.update({'font.size': 22})
plt.scatter(y_test, gbr_pred, s=20)
plt.title('Gradient Boosting Regressor')
plt.xlabel('Actual Crop Production')
plt.ylabel('Predicted Crop Production')
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
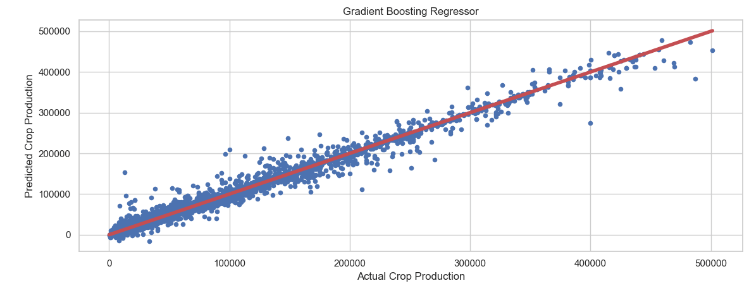
- SGD Regressor + Grid Search CV
from sklearn.linear_model import SGDRegressor
param_grid = {
'learning_rate': ['constant', "optimal", "invscaling", "adaptive" ]
}
sgd = SGDRegressor()
model = GridSearchCV(estimator=sgd, param_grid=param_grid, cv=5,verbose=0)
model.fit(x_train, y_train)
sgd_pred = model.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, sgd_pred)
row = {"Model": "SGDRegressor", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
Regression Report:
Mean Absolute Error : 30368.905
Median Absolute Error : 21651.677
Mean Squared Error : 1848725829.406
Root Mean Squared Error : 42996.812
Max error : 293166.963
R2 score : 0.744
Explained variance score : 0.744
Mean Absolute Percentage Error: 0.961
SGD Regressor: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, sgd_pred, s=20)
plt.title('SGD Regression',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
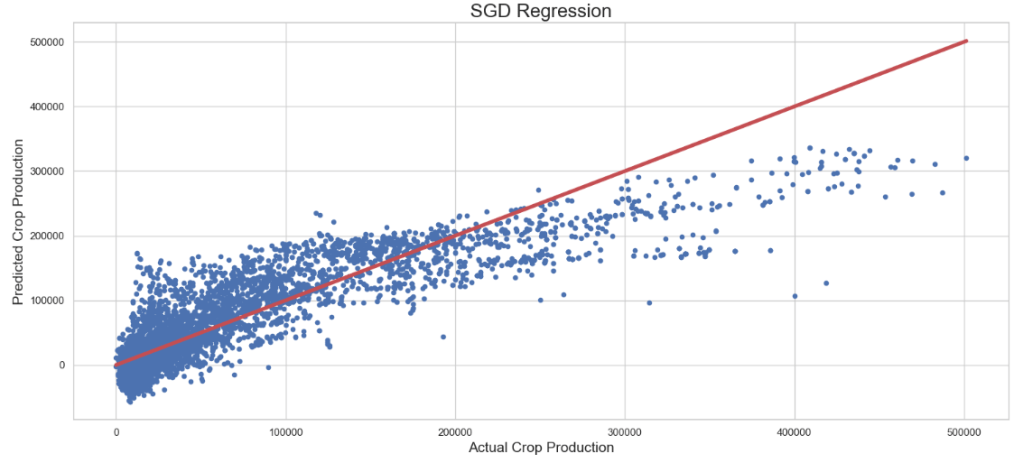
- Ridge Regressor + Grid Search CV
from sklearn.linear_model import Ridge
param_grid = {
'alpha': [0.001, 0.01, 0.1, 1, 2, 3, 4, 5]
}
rid = Ridge()
model = GridSearchCV(estimator=rid, param_grid=param_grid, cv=5, verbose=0)
model.fit(x_train, y_train)
GridSearchCV(cv=5, estimator=Ridge(),
param_grid={'alpha': [0.001, 0.01, 0.1, 1, 2, 3, 4, 5]})
print("The best parameters are %s with a score of %0.2f"
% (model.best_params_, model.best_score_))
rid_pred = model.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, rid_pred)
row = {"Model": "RidgeRegression", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
The best parameters are {'alpha': 5} with a score of 0.75
Regression Report:
Mean Absolute Error : 30366.278
Median Absolute Error : 21672.704
Mean Squared Error : 1848693387.670
Root Mean Squared Error : 42996.435
Max error : 293175.625
R2 score : 0.744
Explained variance score : 0.744
Mean Absolute Percentage Error: 0.961
Ridge Regressor: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, rid_pred, s=20)
plt.title('Ridge Regression',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
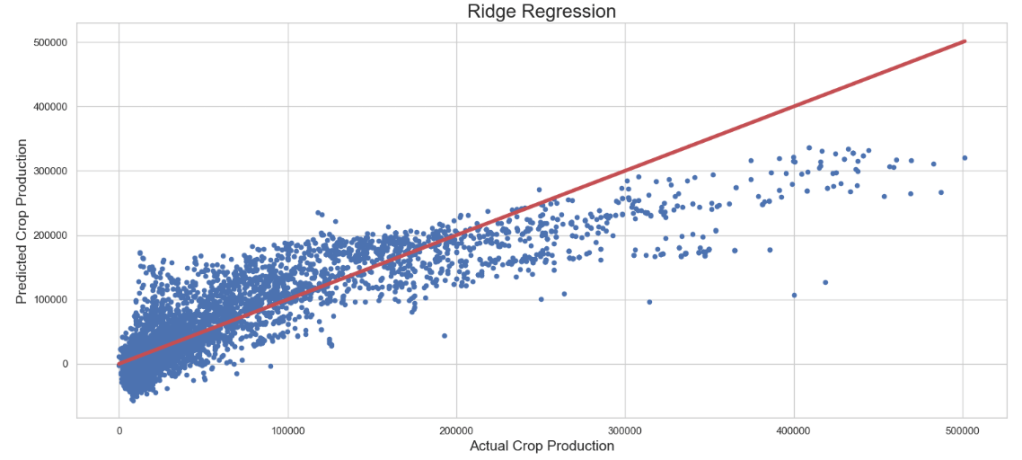
- Lasso Regression
from sklearn.linear_model import Lasso
model = Lasso(alpha= 2, max_iter=10000, tol=0.001)
model.fit(x_train, y_train)
lass_pred = model.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, lass_pred)
row = {"Model": "LassoRegression", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
Regression Report:
Mean Absolute Error : 30365.686
Median Absolute Error : 21654.177
Mean Squared Error : 1848719390.779
Root Mean Squared Error : 42996.737
Max error : 293146.259
R2 score : 0.744
Explained variance score : 0.744
Mean Absolute Percentage Error: 0.961
Lasso Regressor: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, lass_pred, s=20)
plt.title('Lasso Regression',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
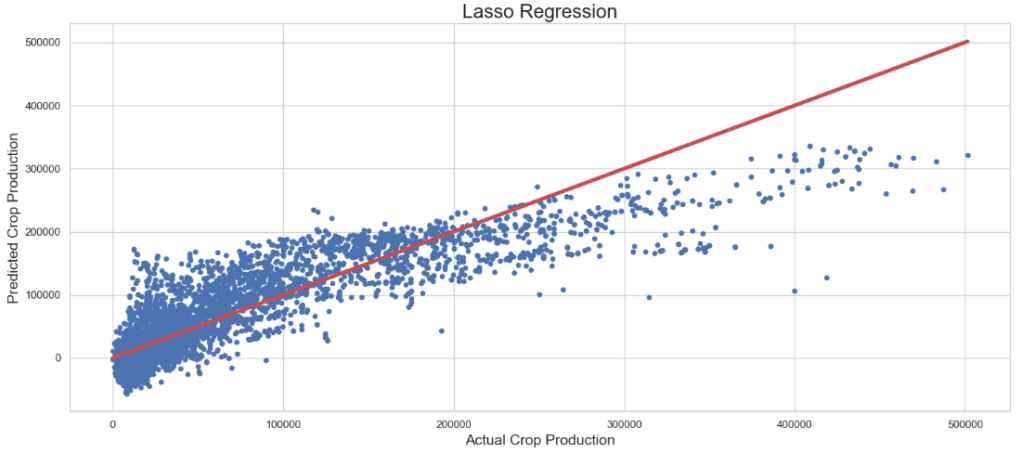
- SVR + Randomized Search CV
from sklearn.svm import SVR
param_dist = {
'kernel' : ['rbf'],
'C' : sp.stats.randint(1, 10001),
"epsilon" : sp.stats.uniform(0.00001, 0.99),
'gamma' : ['scale', 'auto']
}
svr = SVR()
svrcv = RandomizedSearchCV(
estimator=svr,
param_distributions=param_dist,
cv=3,
verbose=0
)
svrcv.fit(x_train, y_train)
print("The best parameters are %s with a score of %0.2f"
% (svrcv.best_params_, svrcv.best_score_))
The best parameters are {'C': 9798, 'epsilon': 0.9537504981825164, 'gamma': 'auto', 'kernel': 'rbf'} with a score of 0.75
svrc_pred = svrcv.predict(x_test)
SVR + Randomized Search CV: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, svrc_pred, s=20)
plt.title('Support Vector Regressor',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
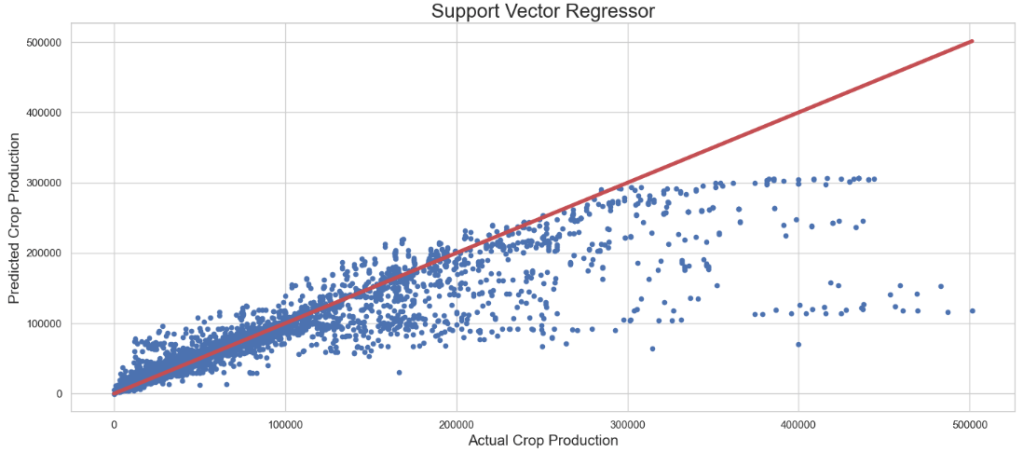
- Elastic Net Regression
from sklearn.linear_model import ElasticNet
model = ElasticNet()
model.fit(x_train, y_train)
en_pred = model.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, en_pred)
row = {"Model": "ElasticNet", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
Regression Report:
Mean Absolute Error : 31848.603
Median Absolute Error : 20473.156
Mean Squared Error : 2373038555.698
Root Mean Squared Error : 48713.844
Max error : 314896.007
R2 score : 0.672
Explained variance score : 0.672
Mean Absolute Percentage Error: 1.082
Elastic Net Regression: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, en_pred, s=20)
plt.title('Elastic Net',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
- Random Forest Regressor + Randomized Search CV
from sklearn.ensemble import RandomForestRegressor
# Create the random grid
random_grid = {'n_estimators': [int(x) for x in np.linspace(start = 100, stop = 1200, num = 12)],
'max_features': ['auto', 'sqrt'],
'max_depth': [int(x) for x in np.linspace(5, 30, num = 6)],
'min_samples_split': [2, 5, 10, 15, 100],
'min_samples_leaf': [1, 2, 5, 10],
'bootstrap' : [True, False]
}
rfr = RandomForestRegressor()
rfrcv = RandomizedSearchCV(
estimator=rfr,
param_distributions=random_grid,
cv=3,
verbose=0
)
rfrcv.fit(x_train, y_train)
RandomizedSearchCV(cv=3, estimator=RandomForestRegressor(),
param_distributions={'bootstrap': [True, False],
'max_depth': [5, 10, 15, 20, 25, 30],
'max_features': ['auto', 'sqrt'],
'min_samples_leaf': [1, 2, 5, 10],
'min_samples_split': [2, 5, 10, 15,
100],
'n_estimators': [100, 200, 300, 400,
500, 600, 700, 800,
900, 1000, 1100,
1200]})
print("The best parameters are %s with a score of %0.2f"
% (rfrcv.best_params_, rfrcv.best_score_))
rfr_pred = rfrcv.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, rfr_pred)
row = {"Model": "RandomForestRegressor", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
The best parameters are {'n_estimators': 1000, 'min_samples_split': 100, 'min_samples_leaf': 1, 'max_features': 'sqrt', 'max_depth': 30, 'bootstrap': False} with a score of 0.92
Regression Report:
Mean Absolute Error : 12283.925
Median Absolute Error : 6875.181
Mean Squared Error : 427473740.868
Root Mean Squared Error : 20675.438
Max error : 239840.455
R2 score : 0.941
Explained variance score : 0.941
Mean Absolute Percentage Error: 0.422
Random Forest Regressor + Randomized Search CV: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, rfr_pred, s=20)
plt.title('Random Forest Regressor',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()
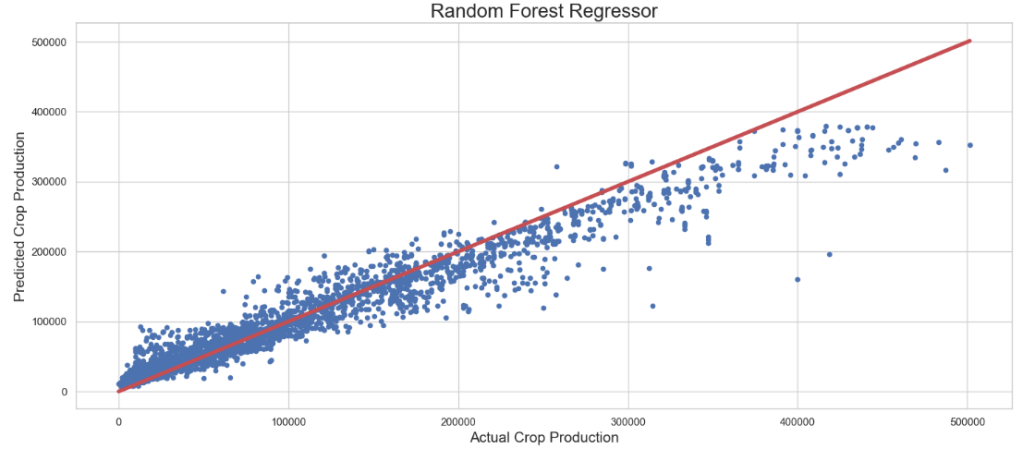
- XGB Regressor + HPO
from xgboost import XGBRegressor
from hyperopt import hp, fmin, tpe, STATUS_OK, Trials
space={'max_depth': hp.quniform("max_depth", 3, 18, 1),
'learning_rate': hp.loguniform('learning_rate', np.log(0.001), np.log(0.5)),
'gamma': hp.uniform ('gamma', 1,9),
'reg_alpha' : hp.uniform('reg_alpha', 0.001, 1.0),
'reg_lambda' : hp.uniform('reg_lambda', 0,1),
'colsample_bytree' : hp.uniform('colsample_bytree', 0.5,1),
'min_child_weight' : hp.quniform('min_child_weight', 0, 10, 1),
'n_estimators': hp.uniform("n_estimators", 100, 20001)
}
def objective(space):
rego=XGBRegressor(
n_estimators =int(space['n_estimators']), max_depth = int(space['max_depth']),
learning_rate = space['learning_rate'], gamma = space['gamma'],
reg_alpha = space['reg_alpha'], min_child_weight=int(space['min_child_weight']),
reg_lambda = space['reg_lambda'],
colsample_bytree=int(space['colsample_bytree']), eval_metric="rmse",
early_stopping_rounds=10)
evaluation = [( x_train, y_train), ( x_test, y_test)]
rego.fit(x_train, y_train,
eval_set=evaluation, verbose=0)
y_pred = rego.predict(x_test)
r_squared = r2_score(y_test, y_pred)
return {'loss': -r_squared, 'status': STATUS_OK }
trials = Trials()
best_hyperparams = fmin(fn = objective,
space = space,
algo = tpe.suggest,
max_evals = 100,
trials = trials)
best_hyperparams['max_depth'],best_hyperparams['n_estimators'] = int(best_hyperparams['max_depth']),int(best_hyperparams['n_estimators'])
model = XGBRegressor(**best_hyperparams)
model.fit(x_train, y_train)
xgb_pred = model.predict(x_test)
mae, medae, mse, rmse, maxerr, r_squared, evs, mape = regression_report(y_test, xgb_pred)
row = {"Model": "XGBRegressor", "MAE": mae,"MEDAE": medae, "MSE": mse, "RMSE": rmse,
"Max Error": maxerr, "R2 Score": r_squared, "EVS": evs, "MAPE": mape}
df_models = df_models.append(row, ignore_index=True)
Regression Report:
Mean Absolute Error : 7784.185
Median Absolute Error : 4403.359
Mean Squared Error : 177196950.303
Root Mean Squared Error : 13311.534
Max error : 213066.703
R2 score : 0.975
Explained variance score : 0.976
Mean Absolute Percentage Error: 0.237
XGB Regressor + HPO: Actual Crop Production vs Predicted Crop Production
plt.figure(figsize=(15,7))
plt.scatter(y_test, xgb_pred, s=20)
plt.title('XGB Regressor',fontsize=20)
plt.xlabel('Actual Crop Production',fontsize=15)
plt.ylabel('Predicted Crop Production',fontsize=15)
plt.plot([min(y_test), max(y_test)], [min(y_test), max(y_test)], color='r', linewidth = 4)
plt.tight_layout()

- QC Comparison of 7 Regression Models
df_models.sort_values(by="R2 Score")

R2-Score of 7 Regression Models
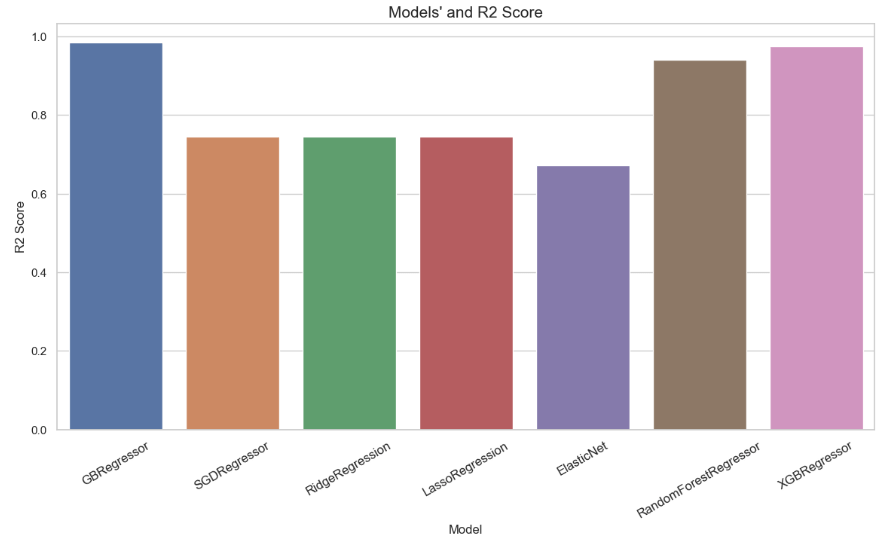
RMSE of Regression Models
plt.figure(figsize=(20,7))
sns.lineplot(x=df_models.Model, y=df_models.RMSE,lw=4)
plt.title("Models' and RMSE ", size=15)
plt.xticks(rotation=30, size=12)
plt.show()
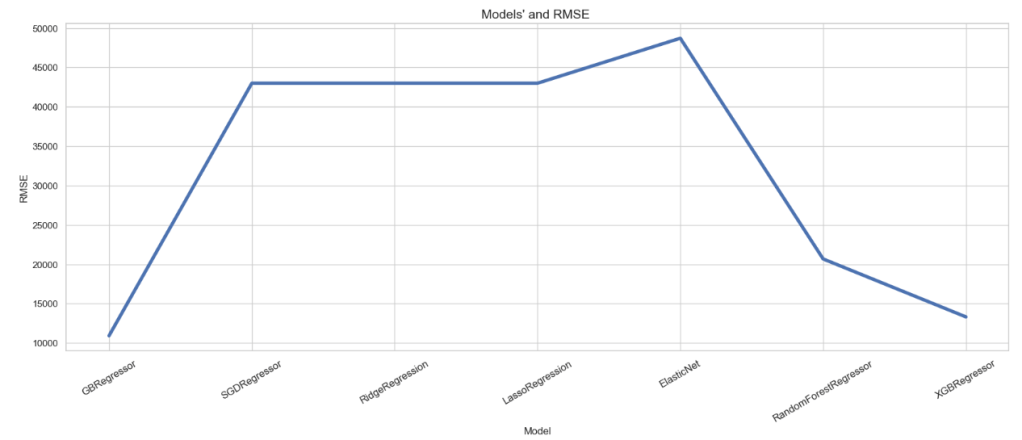
Summary
- We have addressed the ML-based crop yield prediction. This is an essential predictive analytics technique in the agriculture industry. It is an agricultural practice that can help farmers and farming businesses predict crop yield in a particular season when to plant a crop, and when to harvest for better crop yield.
- Several ML algorithms have been applied to support crop yield prediction studies. We have performed a systematic QC comparison of these algorithms using 4 public-domain datasets (courtesy of FAO and World Data Bank).
- CatBoost/SHAP Indian Crop Yield: top SHAP value for log10 yield is Coconut, CatBoost Regressor RMSE score for train is 0.152 dex and for test is 0.168 dex, top season is summer, and top state is Delphi.
- ML-Powered Indian Agriculture:
| Train/Test Data | Algorithm | R2-Score Train | R2-Score Test |
| After Transform + VIF | Random Forest Regressor | 0.99 | 0.95 |
- Global Crop Yield Prediction: the predicted-Actual Yield Test data X-plot is reasonable for the Decision tree algorithm; top yield predictions are potatoes, sweet potatoes, and cassava.
- Crop Recommendation: Multi-label classification of 21 crops leads to severe data overfitting ; Naïve Bayes and RF are the best performers in terms of Accuracy.
- Effects of Climate Changes: Best performers are XGB and GB regressors with HPO and R2-scores of 0.97 and 0.98, respectively.
- However, ML crop yield forecasting is still challenging due to the many factors involved, such as crop and variety, soil type, management practices, pests and diseases, and climate and weather patterns during the season.
- The Road Ahead: Crop Yield Prediction Using ML And Flask Deployment.
Explore More
Make a one-time donation
Make a monthly donation
Make a yearly donation
Choose an amount
Or enter a custom amount
Your contribution is appreciated.
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthlyDonate yearly

Leave a comment