- The Kalman Filter (KF) is a robust tool for creating the optimal solution to many tracking, navigation, and data prediction tasks.
- This article outlines a simplified implementation of KF with commonly used Python commands.
- The main purpose of proposing a tutorial like this is to allow readers clearly understand KF without much knowledge in numerical optimization methods and digital signal processing.
- KF is an optimal state estimator under the assumption of linearity and Gaussian noise. It considers that all measured data have a certain error compared to the actual states. By comparing the next observed state and the predicted state, the predicted model can be iteratively updated.
- KF minimizes posterior state estimation, which is equivalent to saying it minimizes mean-square error estimator.
- The most important concepts are that KF are discrete in time and recursive. In addition, KF predict the future by deriving an adjusted estimate of a state from both prediction and measurements.
- The basic 1D Kalman filter algorithm is implemented as follows:
- Input the measurement vector z as a function of states and
- Input the RMS deviation of sensor data noise R;
- Define the initial state x(k)=0 and P(k)=1, where k=0;
- Set k=k+1 and define the state prediction x*(k)=x(k-1) and the prior error covariance P*(k)=P(k-1);
- Compute the Kalman gain K(k) = P*(k)/(P*(k)+R)
- Update the error covariance P(k) = {1-K(k)} x P*(k)
- Correct the state x(k) = x*(k) + K(k) x {z(k)-x*(k)}
- Let’s look at several simple illustrative examples of using KF aimed at building intuition and experience, not formal proofs.
Table of Contents
- A Simple 1D Voltage Reading
- Linear True State & Random Noise
- 1D Object Tracking
- 1D State Space Models and Kalman Filtering
- KO Log Returns
- Minimal 1D Kalman Filter
- 1D Kalman Smoothing
- 1D Kalman Time-Series Prediction
- Summary
- References
A Simple 1D Voltage Reading
- Following the quick guide, let’s consider a scalar constant voltage reading a V with the standard deviation of noisy readings of R=0.1 V:
import matplotlib.pyplot as plt
ys = [0.39,0.50,0.48,0.29,0.25,0.32,0.34,0.48,0.41,0.45]
xs = [x for x in range(len(ys))]
plt.figure(figsize=(10,6))
plt.rcParams.update({'font.size': 16})
plt.plot(xs, ys,lw=4)
plt.xlabel('Time (ms)')
plt.ylabel('Value (V)')
plt.grid()
plt.show()
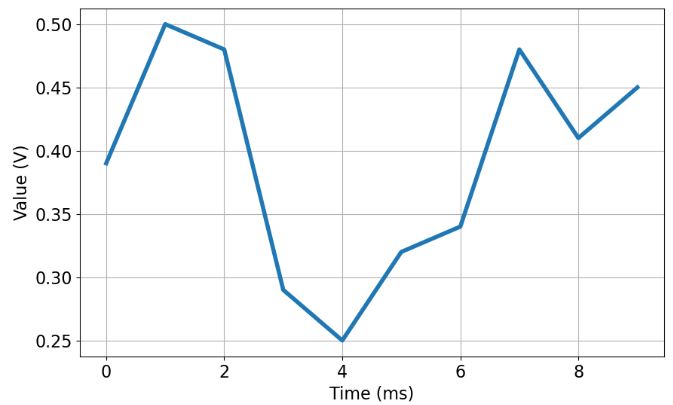
- Defining the initial state parameters
pvec = [1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0]
yk = [0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0]
kvec=[0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0]
rpar=0.1
- KF update with k=0
kvec[0]=pvec[0]/(pvec[0]+rpar)
print(kvec[0])
0.9090909090909091
pvec[0]=(1.0-kvec[0])*pvec[0]
print(pvec[0])
0.09090909090909094
yk[0]=yk[0]+kvec[0]*(ys[0]-yk[0])
print(yk[0])
0.35454545454545455
- KF update with k=1
kvec[1]=pvec[0]/(pvec[0]+rpar)
print(kvec[1])
0.4761904761904763
pvec[1]=(1.0-kvec[1])*pvec[0]
print(pvec[1])
0.04761904761904763
yk[1]=yk[0]+kvec[1]*(ys[1]-yk[0])
print(yk[1])
0.42380952380952386
- KF update with k=2
kvec[2]=pvec[1]/(pvec[1]+rpar)
print(kvec[2])
0.3225806451612903
pvec[2]=(1.0-kvec[2])*pvec[1]
print(pvec[2])
0.032258064516129045
yk[2]=yk[1]+kvec[2]*(ys[2]-yk[1])
print(yk[2])
0.44193548387096776
- KF update with k=3
kvec[3]=pvec[2]/(pvec[2]+rpar)
print(kvec[3])
0.24390243902439032
pvec[3]=(1.0-kvec[3])*pvec[2]
print(pvec[3])
0.024390243902439032
yk[3]=yk[2]+kvec[3]*(ys[3]-yk[2])
print(yk[3])
0.40487804878048783
- KF update with k=4
kvec[4]=pvec[3]/(pvec[3]+rpar)
print(kvec[4])
0.19607843137254907
pvec[4]=(1.0-kvec[4])*pvec[3]
print(pvec[4])
0.019607843137254905
yk[4]=yk[3]+kvec[4]*(ys[4]-yk[3])
print(yk[4])
0.37450980392156863
- KF update with k=5
k=5
kvec[k]=pvec[k-1]/(pvec[k-1]+rpar)
print(kvec[k])
0.1639344262295082
pvec[k]=(1.0-kvec[k])*pvec[k-1]
print(pvec[k])
0.016393442622950824
yk[k]=yk[k-1]+kvec[k]*(ys[k]-yk[k-1])
print(yk[k])
0.3655737704918033
- KF update with k=6
k=6
kvec[k]=pvec[k-1]/(pvec[k-1]+rpar)
print(kvec[k])
0.14084507042253525
pvec[k]=(1.0-kvec[k])*pvec[k-1]
print(pvec[k])
0.014084507042253525
yk[k]=yk[k-1]+kvec[k]*(ys[k]-yk[k-1])
print(yk[k])
0.3619718309859155
- KF update with k=7
k=7
kvec[k]=pvec[k-1]/(pvec[k-1]+rpar)
print(kvec[k])
0.12345679012345681
pvec[k]=(1.0-kvec[k])*pvec[k-1]
print(pvec[k])
0.012345679012345682
yk[k]=yk[k-1]+kvec[k]*(ys[k]-yk[k-1])
print(yk[k])
0.3765432098765432
- KF update with k=8
k=8
kvec[k]=pvec[k-1]/(pvec[k-1]+rpar)
print(kvec[k])
0.10989010989010992
pvec[k]=(1.0-kvec[k])*pvec[k-1]
print(pvec[k])
0.010989010989010992
yk[k]=yk[k-1]+kvec[k]*(ys[k]-yk[k-1])
print(yk[k])
0.3802197802197802
- KF update with k=9
k=9
kvec[k]=pvec[k-1]/(pvec[k-1]+rpar)
print(kvec[k])
0.09900990099009903
pvec[k]=(1.0-kvec[k])*pvec[k-1]
print(pvec[k])
0.009900990099009903
yk[k]=yk[k-1]+kvec[k]*(ys[k]-yk[k-1])
print(yk[k])
0.38712871287128714
- Plotting observed values vs KF (predicted/corrected) updates
plt.figure(figsize=(10,6))
plt.rcParams.update({'font.size': 16})
plt.plot(xs, ys,label='Observed',lw=4)
plt.plot(xs, yk,label='Kalman Filter',lw=4)
ax.legend(handles=[line1, line2])
plt.xlabel('Time (ms)')
plt.ylabel('Value (V)')
plt.legend(loc="lower right")
plt.grid()
plt.show()
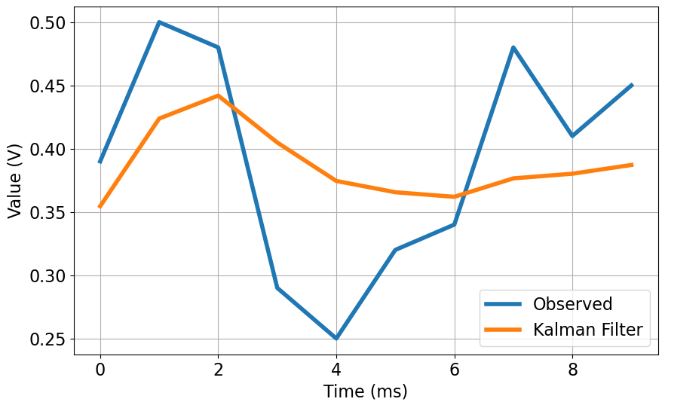
- This plot shows that KF converges to the true voltage value over 8 iterations
import statistics
ykmean=statistics.mean(yk)
print(ykmean)
0.3871115619373332
- The KF prediction error is within the confidence interval
ykstd=statistics.stdev(yk)
print(ykstd)
0.028171057014034436
- Indeed, we can see that
ysmean=statistics.mean(ys)
print(ysmean)
0.391
ysstd=statistics.stdev(ys)
print(ysstd)
0.08774331250237187
Linear True State & Random Noise
- Let’s consider a 1D linear true (simulated) state and measurements with random noise. The KF is expressed in terms of the process and measurement noise covariances Q and R, respectively. Our goal is to examine the suitability of the basic KF algorithm for these measurements:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation, PillowWriter
#Animation to plot incrementally for each time step
def animate(i):
#Clear previous plot
ax.clear()
#Plot to desired time step
plt.plot(x[:i+1], true_states[0][:i+1], label='True State', markersize=10, color='blue')
plt.plot(x[:i+1], measurements[:i+1], 'rx', label='Measurements', markersize=10, color='red')
plt.plot(x[:i+1], filtered_states[0][:i+1], label='Filtered State', color='green')
plt.title('Kalman Filter Position Q=1.0 R=1.0')
plt.xlabel('Time Steps')
plt.ylabel('Position')
plt.legend()
# Kalman filter parameters
dt = 1.0 # Time step
A = np.array([[1, dt], [0, 1]]) # State transition matrix
H = np.array([[1, 0]]) # Measurement matrix
Q = 1.0* np.eye(2) # Process noise covariance
R = 0.1 # Measurement noise covariance
# Initial state estimate
x_hat = np.array([[0], [0]])
# Initial covariance estimate
P = np.eye(2)
# Number of time steps
num_steps = 50
x = np.arange(num_steps)
# True state (simulated)
true_states = np.linspace(0, 2 * np.pi, num_steps)
true_positions = np.cos(true_states)
true_states = np.vstack((true_states, true_positions))
# Measurements with noise
measurements = true_states[0, :] + np.sqrt(R) * np.random.randn(num_steps)
# Kalman filter loop
filtered_states = np.zeros((2, num_steps))
for k in range(num_steps):
# Prediction step
x_hat_minus = A.dot(x_hat)
P_minus = A.dot(P).dot(A.T) + Q
# Update step
K = P_minus.dot(H.T).dot(np.linalg.inv(H.dot(P_minus).dot(H.T) + R))
x_hat = x_hat_minus + K.dot(measurements[k] - H.dot(x_hat_minus))
P = (np.eye(2) - K.dot(H)).dot(P_minus)
filtered_states[:, k] = x_hat.flatten()
# Create a figure and axis
fig, ax = plt.subplots()
#Save animation as gif
animation = FuncAnimation(fig, animate, frames=num_steps, interval=400, repeat=False)
animation.save('kalman_filter_animation.gif', writer='pillow', fps=5)
plt.show()
- Plotting the KF position vs time steps for different values of noise covariances

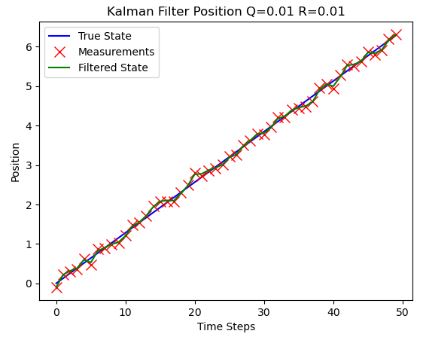


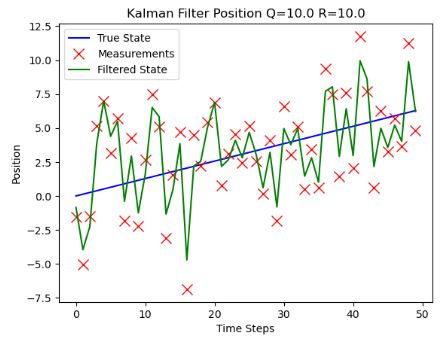
- As expected, robustness of KF with respect to both noise covariances is observed.
1D Object Tracking
- Let’s apply the KF in tracking an object in a 1-D direction using Python. In doing so, we introduce the following class
class KalmanFilter(object):
def __init__(self, dt, u, std_acc, std_meas):
self.dt = dt
self.u = u
self.std_acc = std_acc
self.A = np.matrix([[1, self.dt],
[0, 1]])
self.B = np.matrix([[(self.dt**2)/2], [self.dt]])
self.H = np.matrix([[1,0]])
self.Q = np.matrix([[(self.dt**4)/4, (self.dt**3)/2],
[(self.dt**3)/2, self.dt**2]]) * self.std_acc**2
self.R = std_meas**2
self.P = np.eye(self.A.shape[1])
self.x = np.matrix([[0],[0]])
def predict(self):
# Ref :Eq.(9) and Eq.(10)
# Update time state
self.x = np.dot(self.A, self.x) + np.dot(self.B, self.u)
# Calculate error covariance
# P= A*P*A' + Q
self.P = np.dot(np.dot(self.A, self.P), self.A.T) + self.Q
return self.x
def update(self, z):
# Ref :Eq.(11) , Eq.(11) and Eq.(13)
# S = H*P*H'+R
S = np.dot(self.H, np.dot(self.P, self.H.T)) + self.R
# Calculate the Kalman Gain
# K = P * H'* inv(H*P*H'+R)
K = np.dot(np.dot(self.P, self.H.T), np.linalg.inv(S)) # Eq.(11)
self.x = np.round(self.x + np.dot(K, (z - np.dot(self.H, self.x)))) # Eq.(12)
I = np.eye(self.H.shape[1])
self.P = (I - (K * self.H)) * self.P # Eq.(13)
- The main function defines a model track and creates the KF object by plotting the result of KF predictions vs measurements
def main():
dt = 0.1
t = np.arange(0, 100, dt)
# Define a model track
real_track = 0.1*((t**2) - t)
u= 2
std_acc = 0.25 # we assume that the standard deviation of the acceleration is 0.25 (m/s^2)
std_meas = 1.2 # and standard deviation of the measurement is 1.2 (m)
# create KalmanFilter object
kf = KalmanFilter(dt, u, std_acc, std_meas)
predictions = []
measurements = []
for x in real_track:
# Mesurement
z = kf.H * x + np.random.normal(0, 50)
measurements.append(z.item(0))
predictions.append(kf.predict()[0])
kf.update(z.item(0))
fig = plt.figure()
fig.suptitle('Example of Kalman filter for tracking a moving object in 1-D', fontsize=20)
plt.plot(t, measurements, label='Measurements', color='b',linewidth=0.5)
plt.plot(t, np.array(real_track), label='Real Track', color='y', linewidth=1.5)
plt.plot(t, np.squeeze(predictions), label='Kalman Filter Prediction', color='r', linewidth=1.5)
plt.xlabel('Time (s)', fontsize=20)
plt.ylabel('Position (m)', fontsize=20)
plt.legend()
plt.show()
- Plotting the KF predictions vs measurements and real track
if __name__ == '__main__':
main()

- The above figure shows the simulation performance of 1D KF for the above case. We can see that it handles noisy measurements pretty well. Specifically, KF can help handle random noise or outliers by reducing the impact of individual data points.
- Read more about the aforementioned KF implementation here.
1D State Space Models and Kalman Filtering
- To demonstrate the implementation of 1D state space models and KF, let’s generate some synthetic data with Gaussian noise:
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(0)
# Define the number of time steps
T = 200
# Define the state transition matrix
F = np.array([[1]])
# Define the observation matrix
H = np.array([[1]])
# Define the process noise covariance
Q = np.array([[0.05]])
# Define the observation noise covariance
R = np.array([[1]])
# Define the initial state
x0 = np.array([0])
# Generate the true state and observations
x_true = np.zeros((T, 1))
y = np.zeros(T)
x_true[0] = x0
y[0] = H @ np.asarray(x_true[0]) + np.random.multivariate_normal(np.zeros((1,)), R)
for t in range(1, T):
x_true[t] = F @ x_true[t-1] + np.random.multivariate_normal(np.zeros((1,)), Q)
y[t] = H @ np.asarray(x_true[t]) + np.random.multivariate_normal(np.zeros((1,)), R)
# Initialize the state estimate and covariance
x_hat = np.zeros((T, 1))
P = np.zeros((T, 1, 1))
x_hat[0] = x0
P[0] = Q
# Run the Kalman filter
for t in range(1, T):
# Prediction step
x_hat[t] = F @ x_hat[t-1]
P[t] = F @ P[t-1] @ F.T + Q
# Update step
K = P[t] @ H.T @ np.linalg.inv(H @ P[t] @ H.T + R)
x_hat[t] = x_hat[t] + K @ (y[t] - H @ x_hat[t])
P[t] = (np.eye(1) - K @ H) @ P[t]
- Plotting the true states, observations and estimated states
# Plot the true states, observations and estimated states
plt.figure(figsize=(10, 6))
plt.rcParams.update({'font.size': 18})
plt.plot(range(T), x_true, label='True States',lw=4,color='red')
plt.plot(range(T), y, label='Observations',lw=4,color='orange')
plt.plot(range(T), x_hat, label='Estimated States',lw=4,color='green')
plt.xlabel('Time')
plt.ylabel('State')
plt.legend()
plt.show()
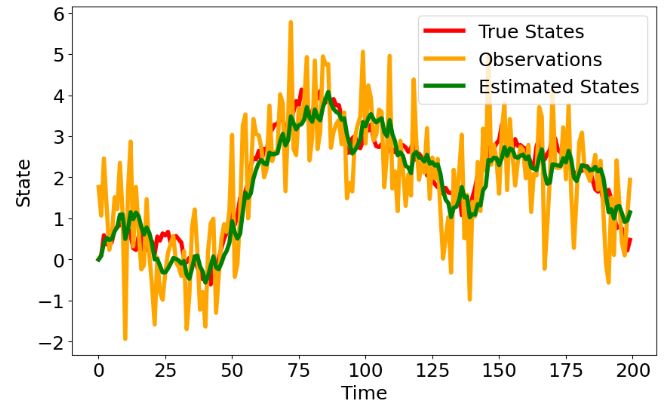
- This test demonstrates that state space models and KF are powerful tools for modeling and estimating the underlying states of a system based on noisy observations.
KO Log Returns
- Let’s apply the above KF technique to the real KO stock price data:
import yfinance as yf
# Download stock price data
data = yf.download('KO', start='2023-01-01', end='2024-01-26')
[*********************100%%**********************] 1 of 1 completed
- Computing log returns and removing missing values
# Compute log returns
data['Log Returns'] = np.log(data['Close']).diff()
# Remove missing values
data = data.dropna()
- Defining input KF parameters, initializing and running KF
# Define the state transition matrix
F = np.array([[1]])
# Define the observation matrix
H = np.array([[1]])
# Define the process noise covariance
Q = np.array([[0.1]])
# Define the observation noise covariance
R = np.array([[0.01]])
# Define the initial state
x0 = np.array([0])
# Define the number of time steps
T = len(data)
# Generate the observations
y = data['Log Returns'].values.reshape(-1, 1)
# Initialize the state estimate and covariance
x_hat = np.zeros((T, 1))
P = np.zeros((T, 1, 1))
x_hat[0] = x0
P[0] = Q
# Run the Kalman filter
for t in range(1, T):
# Prediction step
x_hat[t] = F @ x_hat[t-1]
P[t] = F @ P[t-1] @ F.T + Q
# Update step
K = P[t] @ H.T @ np.linalg.inv(H @ P[t] @ H.T + R)
x_hat[t] = x_hat[t] + K @ (y[t] - H @ x_hat[t])
P[t] = (np.eye(1) - K @ H) @ P[t]
- Plotting the true log returns, estimated log returns and predicted stock prices
# Compute the estimated log returns
log_returns_hat = x_hat.flatten()
# Compute the predicted stock prices
prices_hat = np.exp(np.cumsum(log_returns_hat))
prices = np.exp(np.cumsum(data['Log Returns']))
# Plot the true log returns, estimated log returns and predicted stock prices
plt.figure(figsize=(10, 6))
plt.plot(data.index, data['Log Returns'], label='True Log Returns')
plt.plot(data.index, log_returns_hat, label='Estimated Log Returns')
#plt.plot(data.index, prices_hat, label='Predicted Prices')
plt.xlabel('Date')
plt.ylabel('Log Returns')
plt.legend()
plt.show()
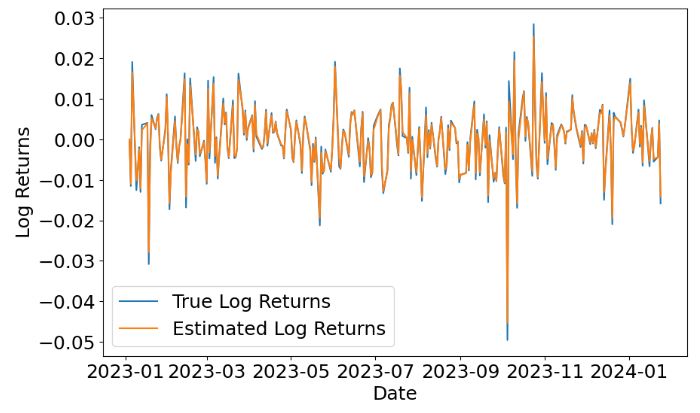
- This plot shows that KF can predict the KO log returns with very reasonable accuracy.
Minimal 1D Kalman Filter
- In this section, we will look at minkf – minimal Kalman filter in Python. This is a minimal implementation with only
Numpydependency. - We generate some 1D random walk data and reconstruct it with KF. The forward and observation models are just identities. The user can either give the model and error covariance matrices as lists, which enable using different values for each time step. If the matrices are given as constant Numpy arrays, the same matrices are used for every time step:
import numpy as np
import minkf as kf
import matplotlib.pyplot as plt
y = np.cumsum(np.random.standard_normal(100))
x0 = np.array([0.0])
Cest0 = 1*np.array([[1.0]])
M = np.array([[1.0]])
K = np.array([[1.0]])
Q = 0.1*np.array([[1.0]])
R = 0.8*np.array([[1.0]])
from IPython.core.display import display, HTML
display(HTML("<style>div.output_scroll { height: 90em; }</style>"))
res = kf.run_filter(y, x0, Cest0, M, K, Q, R, likelihood=True)
res_smo = kf.run_smoother(y, x0, Cest0, M, K, Q, R)
plt.figure(figsize=(10,6))
plt.title('1D Random Walk Data vs Kalman Filter Q=0.1, R=0.8')
plt.rcParams.update({'font.size': 14})
plt.xlabel('Time Step')
plt.plot(y, 'bo', ms=5)
plt.plot(res['x'], 'k-')
plt.plot(res_smo['x'], 'r-')
plt.legend(["Measurements", "True State","Kalman Filter"], loc="lower right")
plt.grid(True)
plt.show()

- Result is a dict that contains the estimated states and the filtering/smoothing covariances
res['loglike']
242.04184667513363
- Sampling from the posterior of the states given all the data can be done via the
samplefunction. Sampling needs the KF results, the dynamics model matrix and model error covariance:
samps = kf.sample(res, M, Q, nsamples=10)
from IPython.core.display import display, HTML
display(HTML("<style>div.output_scroll { height: 90em; }</style>"))
plt.figure(figsize=(10,6))
plt.rcParams.update({'font.size': 14})
plt.title('Sampling from the Posterior of the States')
plt.xlabel('Time Step')
plt.plot(y, 'bo', ms=2)
plt.plot(np.array(samps).T, 'r-', alpha=0.2)
plt.legend(["Measurements", "Posterior of the States"], loc="lower right")
plt.grid(True)
plt.show()
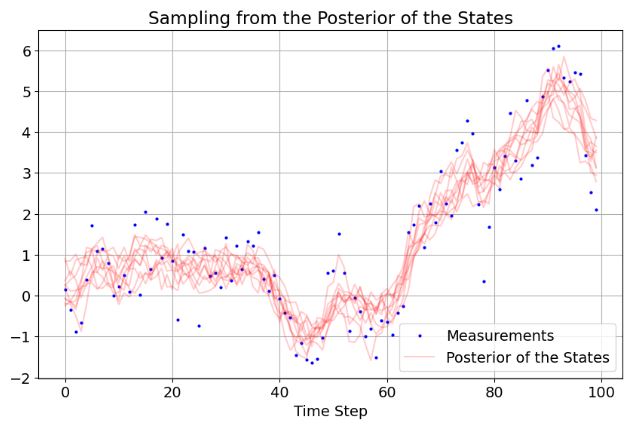
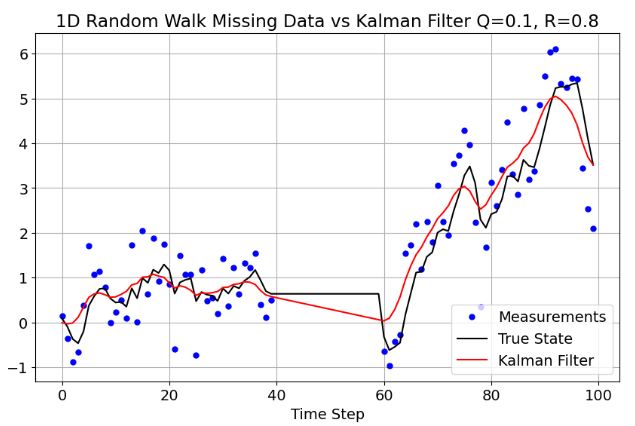
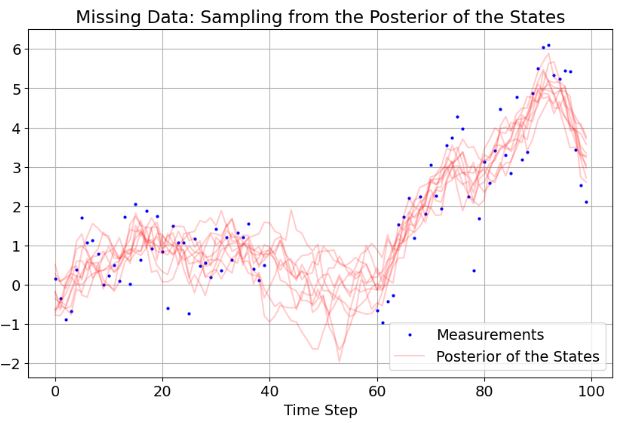
- If any element of the observation vector is
np.nan, the observation is considered missing. Below is the above simple example with some missing observations in the middle:
y_missing = y.copy()
y_missing[40:60] = np.nan
res_missing = kf.run_filter(y_missing, x0, Cest0, M, K, Q, R, likelihood=True)
res_smo_missing = kf.run_smoother(y_missing, x0, Cest0, M, K, Q, R)
from IPython.core.display import display, HTML
display(HTML("<style>div.output_scroll { height: 90em; }</style>"))
plt.figure(figsize=(10,6))
plt.title('1D Random Walk Missing Data vs Kalman Filter Q=0.1, R=0.8')
plt.rcParams.update({'font.size': 14})
plt.xlabel('Time Step')
plt.plot(y_missing, 'bo', ms=5)
plt.plot(res_missing['x'], 'k-')
plt.plot(res_smo_missing['x'], 'r-')
plt.legend(["Measurements", "True State","Kalman Filter"], loc="lower right")
plt.grid(True)
plt.show()
from IPython.core.display import display, HTML
display(HTML("<style>div.output_scroll { height: 90em; }</style>"))
samps_missing = kf.sample(res_missing, M, Q, nsamples=10)
plt.figure(figsize=(10,6))
plt.plot(y_missing, 'bo', ms=2)
plt.plot(np.array(samps_missing).T, 'r-', alpha=0.2)
plt.rcParams.update({'font.size': 14})
plt.xlabel('Time Step')
plt.title('Missing Data: Sampling from the Posterior of the States')
plt.legend(["Measurements", "Posterior of the States"], loc="lower right")
plt.grid(True)
plt.show()
1D Kalman Smoothing
- This section will deal with tsmoothie – a Python library for time-series smoothing and outlier detection in a vectorized way. This library computes, in a fast and efficient way, the smoothing of single or multiple time-series. The smoothing technique available is Kalman Smoothing with customizable components (level, trend, seasonality, and long seasonality).
- In this example, we generate 3 random walk time series of length 100 and apply the Kalman Smoother
import numpy as np
import matplotlib.pyplot as plt
from tsmoothie.smoother import *
from tsmoothie.utils_func import sim_randomwalk
# generate 3 randomwalks timeseries of lenght 100
np.random.seed(123)
data = sim_randomwalk(n_series=3, timesteps=100,
process_noise=10, measure_noise=30)
# operate smoothing
smoother = KalmanSmoother(component='level_trend',
component_noise={'level':0.1, 'trend':0.1})
smoother.smooth(data)
# generate intervals
low, up = smoother.get_intervals('kalman_interval', confidence=0.05)
# plot the first smoothed timeseries with intervals
plt.figure(figsize=(11,6))
plt.plot(smoother.data[0], '.k')
plt.plot(smoother.smooth_data[0], linewidth=3, color='blue')
plt.fill_between(range(len(smoother.data[0])), low[0], up[0], alpha=0.3)
plt.rcParams.update({'font.size': 14})
plt.xlabel('Time Step')
plt.title('Random Walk Data: Kalman Smoothing')
plt.legend(["Measurements", "Kalman Smoother"], loc="lower right")

1D Kalman Time-Series Prediction
- Let’s test the function kalman_filter that estimates hidden states using KF:
import numpy as np
import matplotlib.pyplot as plt
# Function to perform Kalman filtering
def kalman_filter(Y, U, Phi, Ypsilon, A, R, Q, x0, P0):
num_obs = len(Y)
num_hidden = len(x0)
# Initialize arrays to store results
x_pred = np.zeros((num_obs, num_hidden))
P_pred = np.zeros((num_obs, num_hidden, num_hidden))
x_filt = np.zeros((num_obs, num_hidden))
P_filt = np.zeros((num_obs, num_hidden, num_hidden))
x_pred[0] = x0
P_pred[0] = P0
for t in range(1, num_obs):
# Prediction step
x_pred[t] = Ypsilon.T * U[t] + Phi[t] @ x_filt[t - 1]
P_pred[t] = Phi[t] * P_filt[t - 1] * Phi[t].T + Q
# Kalman gain
K = P_pred[t] @ A.T @ np.linalg.inv(A @ P_pred[t] @ A.T + R)
# Update step
x_filt[t] = x_pred[t] + K @ (Y[t] - A @ x_pred[t])
P_filt[t] = P_pred[t] - K @ A @ P_pred[t]
return x_filt, P_filt, x_pred, P_pred
- Consider the example of random 1D data
# Example data
num_obs = 1000
num_hidden = 3
np.random.seed(1)
# Simulate exogenous variable U_t
U = np.random.randn(num_obs).T
- Applying KF to the simulated data and plotting the result
# Specify matrices in model
A = np.array([[1, 1, 0], [0, 0, 1]])
Phi = np.array([np.array([[0, 0, 0], [0, 0.1, 0], [0, -0.1, 0]]) * np.random.standard_normal() for _ in range(num_obs)]) + np.eye(3)
Q = np.array([[1, 0], [0, 1], [-1, -1]])@np.transpose(np.array([[1, 0], [0, 1], [-1, -1]]))
R = np.eye(2) # Assuming the noise in the observed states is uncorrelated
Ypsilon = np.array([[0.2, 0.25, 0.15]]).T
# Simulate variables v, w, X and Y
v = np.random.multivariate_normal(np.zeros(2), R, num_obs)
w = np.random.multivariate_normal(np.zeros(num_hidden), Q, num_obs)
X = np.zeros((num_obs, num_hidden))
for t in range(1, num_obs):
X[t] = (Ypsilon.T * U[t] + Phi[t] @ X[t - 1] + w[t])[0]
Y = np.tensordot(A, X, axes=(1,1)).T + v
# Initial state estimate and covariance
x0 = np.zeros(num_hidden)
P0 = Q.copy()
# Estimate hidden states using Kalman filter
x_filt, p_filt, x_pred, p_pred = kalman_filter(Y, U, Phi, Ypsilon, A, R, Q, x0, P0)
plt.figure(figsize=(10,6))
plt.plot(x_filt[:, 0])
plt.fill_between(range(num_obs), x_filt[:, 0] - 1.96*p_filt[:, 0, 0]**.5, x_filt[:, 0] + 1.96*p_filt[:, 0, 0]**.5, alpha=.4)
plt.plot(X[:, 0])
plt.rcParams.update({'font.size': 14})
plt.xlabel('Time Step')
plt.title('Kalman Prediction of Random Exogenous Data')
plt.legend(['X_predict', 'X_real', 'Confidence bound'])
plt.show()

- This simulation test confirms the validity and effectiveness of using a KF state-space model in 1D time series predictions.
Summary
- Kalman filter (KF) is a powerful algorithm that can be employed in the state estimation problems with low signal-to-noise ratio.
- This post provided a gentle introduction to the 1D KF, a numerical method that is very popular in time series econometrics.
- The aim of the project was to understand the basics of KF in the form of hands-on numerical examples so we could move on to real-world 2D object tracking to be discussed elsewhere.
- In the case of a 1D linear system with measurements errors drawn from a random noise KF has been shown to be the robust estimator.
- The same approach can be implemented to estimate and cancel out the noise of time-variant signals such as stocks, voltage, ECG, GPS, etc. Also, it can be applied to sensor fusion.
- As we saw in our numerical simulations, KF can be incorporated whenever we need to predict the next state of a system based on some noisy measurements.
- In the case if nonlinear systems, the extended KF (EKF) which is a nonlinear version of KF should be considered.
References
- KalmanExample
- Kalman-Filter-Simulation
- 1D-Kalman-Filter
- Optimized-Kalman-Filter
- Object Tracking: Simple Implementation of Kalman Filter in Python
- Advanced Time Series Analysis: State Space Models and Kalman Filtering
One-Time
Monthly
Yearly
Make a one-time donation
Make a monthly donation
Make a yearly donation
Choose an amount
€5.00
€15.00
€100.00
€5.00
€15.00
€100.00
€5.00
€15.00
€100.00
Or enter a custom amount
€
Your contribution is appreciated.
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthlyDonate yearly

Leave a comment