- Breast cancer (BC) is the second leading cause of death in women all over the world and about 12% of women will suffer from this disease during their lifetime.
- Early diagnosis and detection is necessary in order to improve the prognosis of BC.
- In principle, available Machine Learning (ML) and Artificial Intelligence (AI) techniques can extract features from images automatically and then perform BC classification.
- The recently published review shows that the most successful studies related to AI-driven BC detection applied different types of Neural Network (NN) models to a number of publicly available datasets.
- Referring to the earlier AI BC classification experiment and related studies, it appears that one can build and train NN models that accurately predict the probability of BC for individual patients.
- Our goal today is to classify whether the BC is benign or malignant by training and testing a NN-classifier. The final goal to establish an accurate NN model to aid radiologists in BC risk estimation.
Workflow
The entire workflow is as follows:
- Import Key Libraries
- Load Input BC Dataset
- Exploratory Data Analysis (EDA)
- Data Pre-Processing/Editing/Transformation
- Feature Engineering
- Train/Test Data Splitting
- Build TensorFlow/Keras Model
- Compile and Train NN Model
- Examine Training/Validation Accuracy
- Make Predictions on Test data
- Model Classification Report
Prerequisites
We need to install the following libraries:
- Numpy
- Pandas
- Matplotlib
- Seaborn
- Sklearn
- Tensorflow
!pip install –user tensorflow
BC Dataset
In this study, we use the BC Wisconsin (Diagnostic) Dataset to predict whether the BC is benign or malignant.
Model features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass shown below:
img1 = plt.imread(‘malignant.png’)
img2 = plt.imread(‘benign.png’)
fig, (ax, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
fig.subplots_adjust(wspace=0.5)
ax.imshow(img1)
ax.title.set_text(‘Malignant’)
ax2.imshow(img2)
ax2.title.set_text(‘Benign’)
plt.show()

Attribute Information:
1) ID number
2) Diagnosis (M = malignant, B = benign)
3-32)
Ten real-valued features are computed for each cell nucleus:
a) radius (mean of distances from center to points on the perimeter)
b) texture (standard deviation of gray-scale values)
c) perimeter
d) area
e) smoothness (local variation in radius lengths)
f) compactness (perimeter^2 / area – 1.0)
g) concavity (severity of concave portions of the contour)
h) concave points (number of concave portions of the contour)
i) symmetry
j) fractal dimension (“coastline approximation” – 1)
All feature values are recorded with 4 significant digits.
Missing attribute values: none
Class distribution: 357 benign, 212 malignant.
TechVidvan NN Model
Let’s set the working directory YOURPATH
import os
os.chdir(‘YOURPATH’) # Set working directory
os. getcwd()
and import libraries for BC classification
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
Let’s read the input Kaggle data csv file data.csv
df = pd.read_csv(‘data.csv’)
df.head(10)

10 rows × 33 columns
Let’s count the number of rows and columns in dataset:
df.shape
(569, 33)
Let’s look at the pair plot of the input data (selected features)
fig=sns.pairplot(df,hue = ‘diagnosis’, palette= ‘tab10’, vars = [‘radius_mean’, ‘texture_mean’, ‘perimeter_mean’,’area_mean’,’smoothness_mean’])
fig.savefig(“bcdpairplot.png”)

Also, we can count the number of empty values in each column
df.isna().sum()
drop the columns with all the missing values:
df = df.dropna(axis = 1)
df.shape
and get the count of the number of Malignant(M) or Benign(B) cells
df[‘diagnosis’].value_counts()
B 357 M 212 Name: diagnosis, dtype: int64
Let’s visualize the count
fig=sns.countplot(df[‘diagnosis’], label = ‘count’)
plt.savefig(“bcdcount.png”)
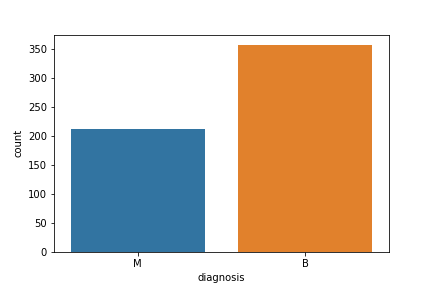
Let’s look at the data types to see which columns need to be encoded:
df.dtypes
id int64 diagnosis object radius_mean float64 texture_mean float64 perimeter_mean float64 area_mean float64 smoothness_mean float64 compactness_mean float64 concavity_mean float64 concave points_mean float64 symmetry_mean float64 fractal_dimension_mean float64 radius_se float64 texture_se float64 perimeter_se float64 area_se float64 smoothness_se float64 compactness_se float64 concavity_se float64 concave points_se float64 symmetry_se float64 fractal_dimension_se float64 radius_worst float64 texture_worst float64 perimeter_worst float64 area_worst float64 smoothness_worst float64 compactness_worst float64 concavity_worst float64 concave points_worst float64 symmetry_worst float64 fractal_dimension_worst float64 dtype: object
Rename the diagnosis data to labels:
df = df.rename(columns = {‘diagnosis’ : ‘label’})
print(df.dtypes)
and define the dependent variable that will be predicted (label)
y = df[‘label’].values
print(np.unique(y))
['B' 'M']
Let’s perform encoding categorical data from text(B and M) to integers (0 and 1)
from sklearn.preprocessing import LabelEncoder
labelencoder = LabelEncoder()
Y = labelencoder.fit_transform(y) # M = 1 and B = 0
print(np.unique(Y))
[0 1]
Let’s define the independent variables (features) by dropping label and id
X = df.drop(labels=[‘label’,’id’],axis = 1)
whilst applying MinMaxScaler() to X
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaler.fit(X)
X = scaler.transform(X)
print(X)
[[0.52103744 0.0226581 0.54598853 ... 0.91202749 0.59846245 0.41886396] [0.64314449 0.27257355 0.61578329 ... 0.63917526 0.23358959 0.22287813] [0.60149557 0.3902604 0.59574321 ... 0.83505155 0.40370589 0.21343303] ... [0.45525108 0.62123774 0.44578813 ... 0.48728522 0.12872068 0.1519087 ] [0.64456434 0.66351031 0.66553797 ... 0.91065292 0.49714173 0.45231536] [0.03686876 0.50152181 0.02853984 ... 0. 0.25744136 0.10068215]]
Let’s split the data into training and testing datasets (with test_size = 0.25) in order to verify the accuracy after fitting the model
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test = train_test_split(X,Y, test_size = 0.25, random_state=42)
print(‘Shape of training data is: ‘, x_train.shape)
print(‘Shape of testing data is: ‘, x_test.shape)
Shape of training data is: (426, 30) Shape of testing data is: (143, 30)
Let’s invoke TensorFlow with Keras optimizers and create the Sequential model
import tensorflow
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation, Dropout
model = Sequential()
model.add(Dense(128, input_dim=30, activation=’relu’))
model.add(Dropout(0.5))
model.add(Dense(64,activation = ‘relu’))
model.add(Dropout(0.5))
model.add(Dense(1))
model.add(Activation(‘sigmoid’))
Let’s compile the NN model
model.compile(loss = ‘binary_crossentropy’, optimizer = ‘adam’ , metrics = [‘accuracy’])
model.summary()
Model: "sequential_4"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_12 (Dense) (None, 128) 3968
dropout_8 (Dropout) (None, 128) 0
dense_13 (Dense) (None, 64) 8256
dropout_9 (Dropout) (None, 64) 0
dense_14 (Dense) (None, 1) 65
activation_4 (Activation) (None, 1) 0
=================================================================
Total params: 12,289
Trainable params: 12,289
Non-trainable params: 0
_________________________________________________________________
let’s fit the model with no early stopping or other callbacks:
history = model.fit(x_train,y_train,verbose = 1,epochs = 100, batch_size = 64,validation_data = (x_test,y_test))
see Appendix A
Let’s plot the training/validation loss vs epochs
loss = history.history[‘loss’]
val_loss = history.history[‘val_loss’]
epochs = range(1,len(loss)+1)
plt.plot(epochs,loss,’y’,label = ‘Training loss’)
plt.plot(epochs,val_loss,’r’,label = ‘Validation loss’)
plt.title(‘TechVidvan NN Training and Validation loss’)
plt.xlabel(‘Epochs’)
plt.ylabel(‘Loss’)
plt.legend()
plt.show()

Let’s plot the training/validation accuracy vs epochs
loss = history.history[‘loss’]
val_loss = history.history[‘val_loss’]
epochs = range(1,len(loss)+1)
plt.plot(epochs,loss,’y’,label = ‘Training loss’)
plt.plot(epochs,val_loss,’r’,label = ‘Validation loss’)
plt.title(‘TechVidvan NN Training and Validation loss’)
plt.xlabel(‘Epochs’)
plt.ylabel(‘Loss’)
plt.legend()
plt.show()

Let’s predict the test set results
y_pred = model.predict(x_test)
y_pred = (y_pred > 0.5)
and plot the confusion matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test,y_pred,normalize=’all’)
sns.heatmap(cm, annot = True)

Let’s look at the binary classification report
from sklearn.metrics import classification_report
target_names = [‘B’, ‘M’]
print(classification_report(y_test, y_pred, target_names=target_names))
precision recall f1-score support
B 0.99 0.98 0.98 89
M 0.96 0.98 0.97 54
accuracy 0.98 143
macro avg 0.98 0.98 0.98 143
weighted avg 0.98 0.98 0.98 143
Let’s look at other NN model evaluation metrics
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_pred, normalize=True)
0.9790209790209791
from sklearn.metrics import cohen_kappa_score
cohen_kappa_score(y_test, y_pred)
0.9555302166476625
from sklearn.metrics import hamming_loss
hamming_loss(y_test, y_pred)
0.02097902097902098
from sklearn.metrics import jaccard_score
jaccard_score(y_test, y_pred)
0.9464285714285714
from sklearn.metrics import log_loss
log_loss(y_test, y_pred)
0.7246008977593886
from sklearn.metrics import matthews_corrcoef
matthews_corrcoef(y_test, y_pred)
0.9556352128340201
Optimized Features + SVM/LR + NN
Let’s optimize the NN model by exploring feature engineering and comparing supervised ML (SVM and LR) against Sequential NN described above.
Let’s set the working directory YOURPATH
import os
os.chdir(‘YOURPATH’) # Set working directory
os. getcwd()
import basic libraries
import pandas as pd
import seaborn as sns # for data visualization
import matplotlib.pyplot as plt # for data visualization
%matplotlib inline
and load the input dataset
Data importing and printing
df = pd.read_csv(‘data.csv’)
print(“\nNot processed data. First 10 rows\n”)
df.head(10)
Not processed data. First 10 rows
as a table 10 rows × 33 columns described above.
Let’s install TensorFlow
!pip install –user tensorflow (
tensorflow-2.10.0-cp39-cp39-win_amd64.whl
)
and import the following key libraries
import tensorflow
import os
import numpy as np
from sklearn.preprocessing import StandardScaler
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
np.random.seed(1337)
os.environ[‘TF_CPP_MIN_LOG_LEVEL’] = ‘3’
pd.set_option(“display.max_column”, 100)
pd.set_option(“display.width”, 1000)
Let’s define the following customized functions
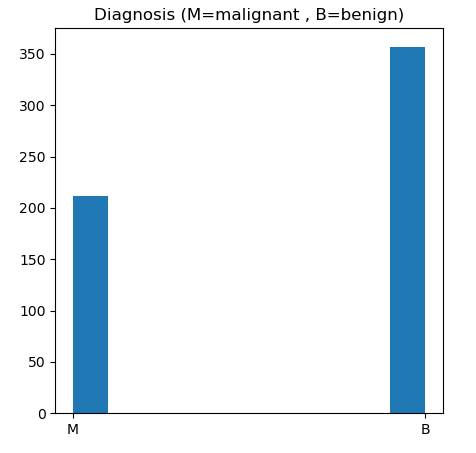
def dataVisualization(data, start=0):
if start == 0:
plt.figure(figsize = (5,5))
plt.hist(data[‘diagnosis’])
plt.title(‘Diagnosis (M=malignant , B=benign)’)
plt.show()
else:
figures(data, start, start+10)
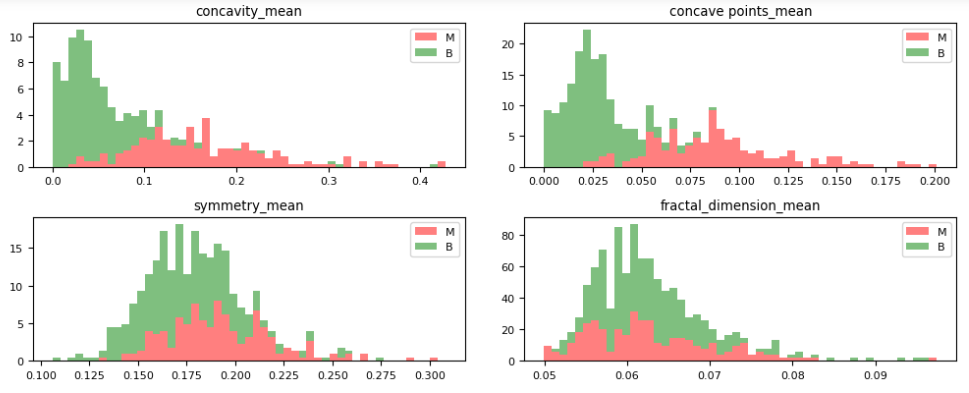
def figures(data, s, e):
plt.rcParams.update({‘font.size’: 8})
fig, axes = plt.subplots(nrows=5, ncols=2, figsize=(10, 10))
axes = axes.ravel()
features = list(data.columns[s:e])
dataM = data[data['diagnosis'] == 'M']
dataB = data[data['diagnosis'] == 'B']
for idx, ax in enumerate(axes):
ax.figure
binwidth = (max(data[features[idx]]) - min(data[features[idx]])) / 50
ax.hist([dataM[features[idx]], dataB[features[idx]]],
bins=np.arange(min(data[features[idx]]), max(data[features[idx]]) + binwidth, binwidth),
alpha=0.5, stacked=True, density=True, label=['M', 'B'], color=['r', 'g'])
ax.legend(loc='upper right')
ax.set_title(features[idx])
plt.tight_layout()
plt.show()
def dataPreparation(df):
data = df.iloc[:, 1:32]
data['diagnosis'] = data['diagnosis'].map(dict(M=int(1), B=int(0)))
data = data.rename(columns={"diagnosis": "cancer"})
data = data.drop(columns=['perimeter_mean', 'area_mean', 'perimeter_se', 'area_se', 'perimeter_worst', 'area_worst'])
scaler = StandardScaler()
data.iloc[:, 1:] = scaler.fit_transform(data.iloc[:, 1:])
return data
def XYSplit(data):
X = data.iloc[:, 1:]
y = data[‘cancer’]
return X, y
def modelCreate():
callback = EarlyStopping(monitor=’val_loss’, patience=17, restore_best_weights=True)
model = Sequential()
model.add(Dense(50, activation='relu', input_dim=24, kernel_regularizer='l2'))
model.add(Dense(20, activation='relu', kernel_regularizer='l2'))
model.add(Dense(1, activation='sigmoid', name='Output'))
model.compile(loss='binary_crossentropy', optimizer='SGD', metrics=['accuracy'])
return model, callback
def trainingPlot(history):
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(history.history['accuracy'], label='accuracy')
ax1.plot(history.history['val_accuracy'], label='val_accuracy')
ax1.set_xlabel('Epoch')
ax1.set_ylabel('Accuracy')
ax1.set_ylim([0.5, 1])
ax1.legend(loc='lower right')
ax2.plot(history.history['loss'], label='loss')
ax2.plot(history.history['val_loss'], label='val_loss')
ax2.set_xlabel('Epoch')
ax2.set_ylabel('Loss')
ax2.legend(loc='lower right')
plt.show()
def confusionMatrix(cmLR, scoreLR, cmSVM, scoreSVM, cmNN, scoreNN):
fig, (ax1, ax2, ax3) = plt.subplots(ncols=3, figsize=(23, 7))
fig.subplots_adjust(wspace=0.1)
sns.heatmap(cmLR, annot=True, fmt=".3f", linewidths=.7, square=True, cmap='Blues_r', ax=ax1,annot_kws={
'fontsize': 16,
'fontweight': 'bold',
'fontfamily': 'serif'})
ax1.set_ylabel('Actual label',fontsize=16)
ax1.set_xlabel('Predicted label',fontsize=16)
all_sample_title = 'Logistic Regression Score: {0}'.format(round(scoreLR, 5))
ax1.title.set_text(all_sample_title)
cbar = ax1.collections[0].colorbar
cbar.ax1.tick_params(labelsize=16)
sns.heatmap(cmSVM, annot=True, fmt=".3f", linewidths=.7, square=True, cmap='Blues_r', ax=ax2,annot_kws={
'fontsize': 16,
'fontweight': 'bold',
'fontfamily': 'serif'})
ax2.set_ylabel('Actual label',fontsize=16)
ax2.set_xlabel('Predicted label',fontsize=16)
all_sample_title = 'Support Vectore Machine Score: {0}'.format(round(scoreSVM, 5))
ax2.title.set_text(all_sample_title)
sns.heatmap(cmNN, annot=True, fmt=".3f", linewidths=.7, square=True, cmap='Blues_r', ax=ax3,annot_kws={
'fontsize': 16,
'fontweight': 'bold',
'fontfamily': 'serif'})
ax3.set_ylabel('Actual label',fontsize=16)
ax3.set_xlabel('Predicted label',fontsize=16)
all_sample_title = 'Neural Network Score: {0}'.format(round(scoreNN, 5))
ax3.title.set_text(all_sample_title)
plt.show()
Let’s plot the B and M images
img1 = plt.imread(‘malignant.png’)
img2 = plt.imread(‘benign.png’)
fig, (ax, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
fig.subplots_adjust(wspace=0.5)
ax.imshow(img1)
ax.title.set_text(‘Malignant’)
ax2.imshow(img2)
ax2.title.set_text(‘Benign’)
plt.show()


We can check the basic descriptive statistics of both BC outcomes
print(‘Diagnosis = B:’)
df[df[‘diagnosis’] == ‘B’].describe()
Diagnosis = B:
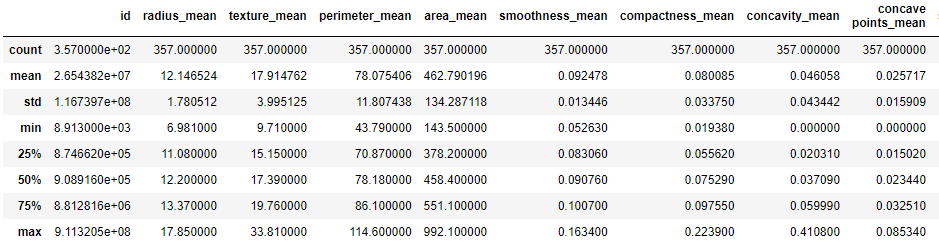
print(‘Diagnosis = M:’)
df[df[‘diagnosis’] == ‘M’].describe()
Diagnosis = M:

Let’s add more columns
new_col = [‘radius_diff’, ‘texture_diff’, ‘perimeter_diff’, ‘area_diff’,
‘smoothness_diff’, ‘compactness_diff’, ‘concavity_diff’,
‘concave points_diff’, ‘symmetry_diff’, ‘fractal_dimension_diff’]
for i, col in zip(range(len(new_col)), new_col):
df[col] = abs(df.iloc[:, i + 2] – df.iloc[:, i + 22])
df.head(10)

let’s plot the BC M vs B outcomes
dataVisualization(df)
Let’s plot the M vs B histograms of our features
dataVisualization(df, 2)
dataVisualization(df, 12)
dataVisualization(df, 22)

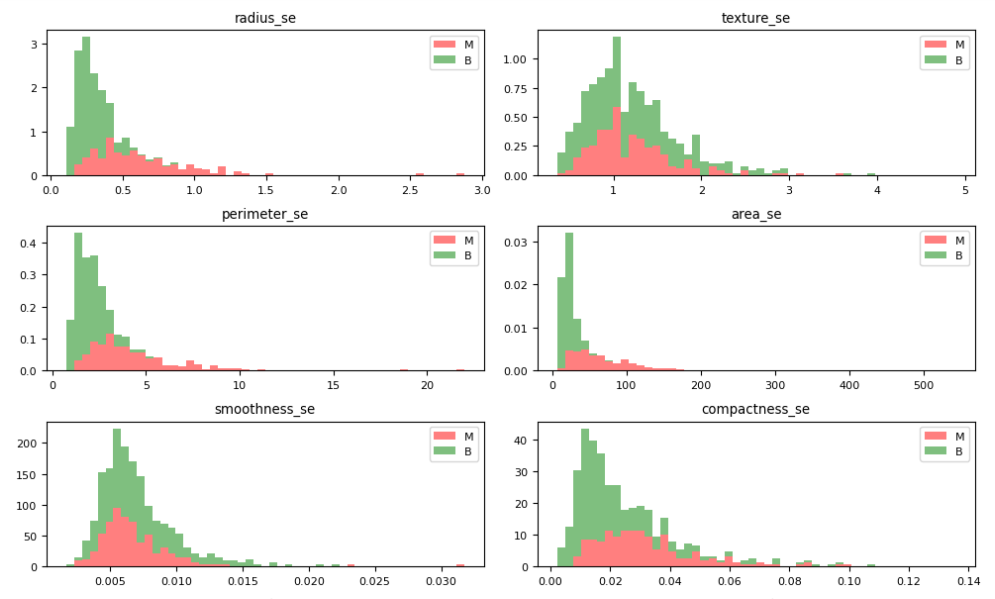

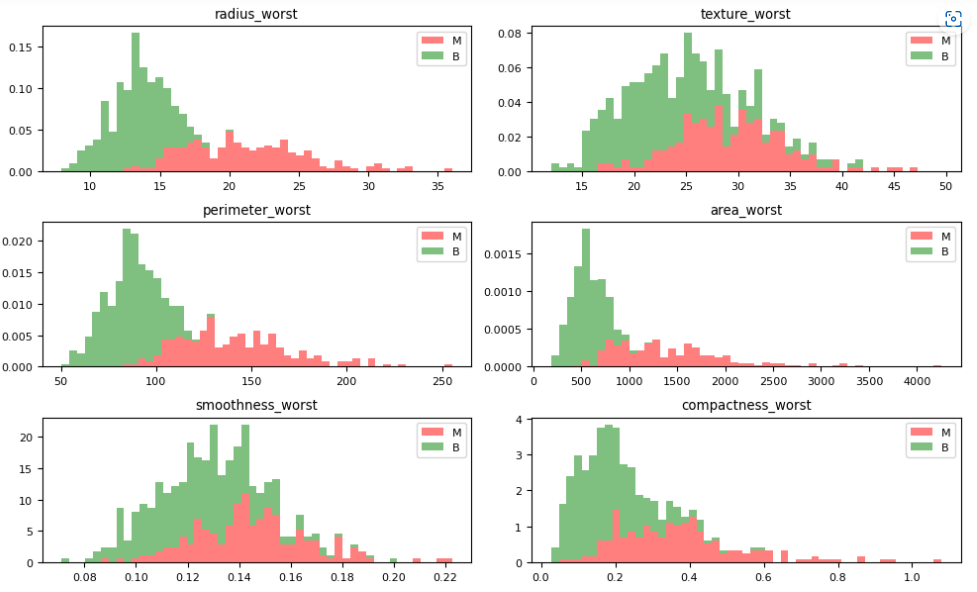

let’s plot the corresponding feature correlation matrices
plt.figure(figsize = (8,8))
corrMatrix = df.iloc[:, 2:12].corr()
sn.heatmap(corrMatrix, annot=True)
plt.show()
plt.figure(figsize = (8,8))
corrMatrix = df.iloc[:, 12:22].corr()
sn.heatmap(corrMatrix, annot=True)
plt.show()
plt.figure(figsize = (8,8))
corrMatrix = df.iloc[:, 22:32].corr()
sn.heatmap(corrMatrix, annot=True)
plt.show()



Data pre-processing:
data = dataPreparation(df)
print(“\nProcessed and standardized data. First 10 rows\n”)
data.head(10)
Processed and standardized data. First 10 rows
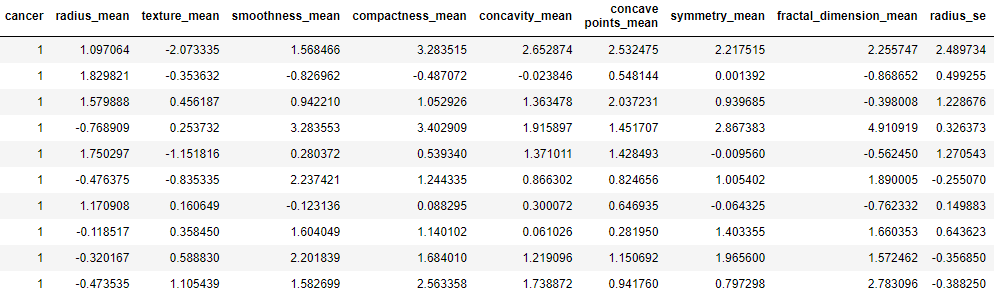
Let’s split the data
from sklearn.model_selection import train_test_split
Split into X and y
X, y = XYSplit(data)
Split into train and test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=7)
print(“The shapes of X_train, X_test, y_train, y_test:”, X_train.shape, X_test.shape, y_train.shape, y_test.shape, ‘\n’)
Let’s compare the Logistic Regression (LR), SVM and NN classifiers
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
Logistic regression training
logreg = LogisticRegression()
logreg.fit(X_train, y_train)
Initialize SVM classifier
SVM = SVC(kernel=’linear’)
SVM.fit(X_train, y_train)
NN model
model, callbacks = modelCreate()
model.summary()
NN training
print(‘\nTraining started…\n’)
history = model.fit(X_train, y_train, epochs=1000, validation_split=0.2, callbacks=[callbacks], verbose=0)
print(f’Training took {len(history.history[“loss”])} epochs’)
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 50) 1250
dense_1 (Dense) (None, 20) 1020
Output (Dense) (None, 1) 21
=================================================================
Total params: 2,291
Trainable params: 2,291
Non-trainable params: 0
_________________________________________________________________
Training started...
Training took 665 epochs
Let’s plot the NN training history
trainingPlot(history)

Let’s make test predictions:
LR testing
scoreLR = logreg.score(X_test, y_test)
SVM testing
scoreSVM = SVM.score(X_test, y_test)
NN testing
scoreNN = model.evaluate(X_test, y_test, verbose=0)[1]
import sklearn
These are LR, SVM, and NN predictions
predictionsLR = logreg.predict(X_test)
cmLR = sklearn.metrics.confusion_matrix(y_test, predictionsLR)
predictionsNN = model.predict(X_test)
predictionsNN = list(map(lambda x: 0 if x < 0.5 else 1, predictionsNN))
cmNN = sklearn.metrics.confusion_matrix(y_test, predictionsNN)
predictionsSVM = SVM.predict(X_test)
cmSVM = sklearn.metrics.confusion_matrix(y_test, predictionsSVM)
And these are the corresponding confusion matrices
confusionMatrix(cmLR, scoreLR, cmSVM, scoreSVM, cmNN, scoreNN)

Let’s look at other model evaluation metrics
print(‘Logistic Regression testing accuracy’, round(scoreLR, 3))
Precision (Accuracy of positive predictions)
precisionLR = cmLR[1,1]/(cmLR[0,1]+cmLR[1,1])
print(‘Precision of Logistic Regression: ‘, round(precisionLR, 3))
Sensitivity=Recall (True Positive Rate)
sensitivityLR = cmLR[1,1]/(cmLR[1,0]+cmLR[1,1])
print(‘Sensitivity/Recall of Logistic Regression: ‘, round(sensitivityLR, 3))
Specificity (True Negative Rate)
specificityLR = cmLR[0,0]/(cmLR[0,0]+cmLR[0,1])
print(‘Specificity of Logistic Regression: ‘, round(specificityLR, 3))
F1-Score (Percent of correct positive predictions)
f1LR = 2(sensitivityLRprecisionLR) / (sensitivityLR+precisionLR)
print(‘F-1 Score of Logistic Regression: ‘, round(specificityLR, 3))
TN FP
FN TP
Logistic Regression testing accuracy 0.972 Precision of Logistic Regression: 1.0 Sensitivity/Recall of Logistic Regression: 0.911 Specificity of Logistic Regression: 1.0 F-1 Score of Logistic Regression: 1.0
print(‘Support Vector Machine testing accuracy’, round(scoreSVM, 3))
Precision (Accuracy of positive predictions)
precisionSVM = cmSVM[1,1]/(cmSVM[0,1]+cmSVM[1,1])
print(‘Precision of Support Vector Machine: ‘, round(precisionSVM, 3))
Sensitivity=Recall (True Positive Rate)
sensitivitySVM = cmSVM[1,1]/(cmSVM[1,0]+cmSVM[1,1])
print(‘Sensitivity/Recall of Support Vector Machine: ‘, round(sensitivitySVM, 3))
Specificity (True Negative Rate)
specificitySVM = cmSVM[0,0]/(cmSVM[0,0]+cmSVM[0,1])
print(‘Specificity of Support Vector Machine: ‘, round(specificitySVM, 3))
F1-Score (Percent of correct positive predictions)
f1SVM = 2(sensitivitySVMprecisionSVM) / (sensitivitySVM+precisionSVM)
print(‘F-1 Score of Support Vector Machine: ‘, round(specificitySVM, 3))
TN FP
FN TP
Support Vector Machine testing accuracy 0.979 Precision of Support Vector Machine: 1.0 Sensitivity/Recall of Support Vector Machine: 0.933 Specificity of Support Vector Machine: 1.0 F-1 Score of Support Vector Machine: 1.0
print(‘Neural Network testing accuracy’, round(scoreNN, 3))
Precision (Accuracy of positive predictions)
precisionNN = cmNN[1,1]/(cmNN[0,1]+cmNN[1,1])
print(‘Precision of Neural Network: ‘, round(precisionNN, 3))
Sensitivity=Recall (True Positive Rate)
sensitivityNN = cmNN[1,1]/(cmNN[1,0]+cmNN[1,1])
print(‘Sensitivity/Recall of Neural Network: ‘, round(sensitivityNN, 3))
Specificity (True Negative Rate)
specificityNN = cmNN[0,0]/(cmNN[0,0]+cmNN[0,1])
print(‘Specificity of Neural Network: ‘, round(specificityNN, 3))
F1-Score (Percent of correct positive predictions)
f1NN = 2(sensitivityNNprecisionNN) / (sensitivityNN+precisionNN)
print(‘F-1 Score of Neural Network: ‘, round(specificityNN, 3))
TN FP
FN TP
Neural Network testing accuracy 0.979 Precision of Neural Network: 1.0 Sensitivity/Recall of Neural Network: 0.933 Specificity of Neural Network: 1.0 F-1 Score of Neural Network: 1.0
Summary
- Our NN model accuracy is 99 % on training data and 98% accuracy on validation data.
- The NN F1-score is 98% for both classes.
- The NN Sensitivity/Recall of Neural Network is 93%.
- Confusion matrices of NN and SVM classifiers are identical.
- High Pair Correlations: radius_mean – area_mean – perimeter_mean, concavity_mean – compactness_mean, and concave_points_mean – perimeter_mean.
Explore More
The Power of AIHealth: Comparison of 12 ML Breast Cancer Classification Models
Supervised ML/AI Breast Cancer Diagnostics (BCD) – The Power of HealthTech
HealthTech ML/AI Q3 ’22 Round-Up
A Comparative Analysis of Breast Cancer ML/AI Binary Classifications
A Comparison of Binary Classifiers for Enhanced ML/AI Breast Cancer Diagnostics – 1. Scikit-Plot
ML/AI Breast Cancer Diagnosis with 98% Confidence
ML/AI Breast Cancer Diagnostics – SciKit-Learn QC
Appendix A
Epoch 1/100 7/7 [==============================] - 0s 17ms/step - loss: 0.6905 - accuracy: 0.5563 - val_loss: 0.6562 - val_accuracy: 0.8252 Epoch 2/100 7/7 [==============================] - 0s 4ms/step - loss: 0.6674 - accuracy: 0.6244 - val_loss: 0.6200 - val_accuracy: 0.8601 Epoch 3/100 7/7 [==============================] - 0s 4ms/step - loss: 0.6362 - accuracy: 0.7324 - val_loss: 0.5793 - val_accuracy: 0.9021 Epoch 4/100 7/7 [==============================] - 0s 4ms/step - loss: 0.5981 - accuracy: 0.7817 - val_loss: 0.5321 - val_accuracy: 0.9161 Epoch 5/100 7/7 [==============================] - 0s 4ms/step - loss: 0.5534 - accuracy: 0.8122 - val_loss: 0.4792 - val_accuracy: 0.9091 Epoch 6/100 7/7 [==============================] - 0s 4ms/step - loss: 0.5106 - accuracy: 0.8216 - val_loss: 0.4238 - val_accuracy: 0.9301 Epoch 7/100 7/7 [==============================] - 0s 4ms/step - loss: 0.4906 - accuracy: 0.8310 - val_loss: 0.3687 - val_accuracy: 0.9441 Epoch 8/100 7/7 [==============================] - 0s 4ms/step - loss: 0.4501 - accuracy: 0.8286 - val_loss: 0.3255 - val_accuracy: 0.9510 Epoch 9/100 7/7 [==============================] - 0s 4ms/step - loss: 0.3961 - accuracy: 0.8779 - val_loss: 0.2818 - val_accuracy: 0.9510 Epoch 10/100 7/7 [==============================] - 0s 4ms/step - loss: 0.3454 - accuracy: 0.8850 - val_loss: 0.2444 - val_accuracy: 0.9510 Epoch 11/100 7/7 [==============================] - 0s 4ms/step - loss: 0.3289 - accuracy: 0.8756 - val_loss: 0.2224 - val_accuracy: 0.9441 Epoch 12/100 7/7 [==============================] - 0s 4ms/step - loss: 0.2757 - accuracy: 0.9155 - val_loss: 0.1917 - val_accuracy: 0.9510 Epoch 13/100 7/7 [==============================] - 0s 3ms/step - loss: 0.2952 - accuracy: 0.8920 - val_loss: 0.1721 - val_accuracy: 0.9441 Epoch 14/100 7/7 [==============================] - 0s 4ms/step - loss: 0.2740 - accuracy: 0.8920 - val_loss: 0.1661 - val_accuracy: 0.9580 Epoch 15/100 7/7 [==============================] - 0s 4ms/step - loss: 0.2697 - accuracy: 0.8991 - val_loss: 0.1570 - val_accuracy: 0.9580 Epoch 16/100 7/7 [==============================] - 0s 3ms/step - loss: 0.2244 - accuracy: 0.9108 - val_loss: 0.1379 - val_accuracy: 0.9441 Epoch 17/100 7/7 [==============================] - 0s 4ms/step - loss: 0.2189 - accuracy: 0.8897 - val_loss: 0.1289 - val_accuracy: 0.9510 Epoch 18/100 7/7 [==============================] - 0s 3ms/step - loss: 0.2353 - accuracy: 0.9085 - val_loss: 0.1317 - val_accuracy: 0.9580 Epoch 19/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1966 - accuracy: 0.9296 - val_loss: 0.1173 - val_accuracy: 0.9720 Epoch 20/100 7/7 [==============================] - 0s 3ms/step - loss: 0.1961 - accuracy: 0.9085 - val_loss: 0.1094 - val_accuracy: 0.9650 Epoch 21/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1926 - accuracy: 0.9061 - val_loss: 0.1066 - val_accuracy: 0.9650 Epoch 22/100 7/7 [==============================] - 0s 3ms/step - loss: 0.1978 - accuracy: 0.9249 - val_loss: 0.1082 - val_accuracy: 0.9720 Epoch 23/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1855 - accuracy: 0.9366 - val_loss: 0.1014 - val_accuracy: 0.9720 Epoch 24/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1641 - accuracy: 0.9507 - val_loss: 0.0922 - val_accuracy: 0.9720 Epoch 25/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1762 - accuracy: 0.9319 - val_loss: 0.0912 - val_accuracy: 0.9790 Epoch 26/100 7/7 [==============================] - 0s 3ms/step - loss: 0.1568 - accuracy: 0.9249 - val_loss: 0.0867 - val_accuracy: 0.9720 Epoch 27/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1681 - accuracy: 0.9319 - val_loss: 0.0850 - val_accuracy: 0.9790 Epoch 28/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1477 - accuracy: 0.9390 - val_loss: 0.0840 - val_accuracy: 0.9790 Epoch 29/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1340 - accuracy: 0.9624 - val_loss: 0.0754 - val_accuracy: 0.9790 Epoch 30/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1570 - accuracy: 0.9437 - val_loss: 0.0815 - val_accuracy: 0.9790 Epoch 31/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1505 - accuracy: 0.9366 - val_loss: 0.0796 - val_accuracy: 0.9790 Epoch 32/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1230 - accuracy: 0.9554 - val_loss: 0.0704 - val_accuracy: 0.9720 Epoch 33/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1404 - accuracy: 0.9484 - val_loss: 0.0700 - val_accuracy: 0.9790 Epoch 34/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1179 - accuracy: 0.9601 - val_loss: 0.0742 - val_accuracy: 0.9790 Epoch 35/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1232 - accuracy: 0.9601 - val_loss: 0.0679 - val_accuracy: 0.9790 Epoch 36/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1211 - accuracy: 0.9624 - val_loss: 0.0655 - val_accuracy: 0.9790 Epoch 37/100 7/7 [==============================] - 0s 5ms/step - loss: 0.1137 - accuracy: 0.9577 - val_loss: 0.0700 - val_accuracy: 0.9790 Epoch 38/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1056 - accuracy: 0.9648 - val_loss: 0.0629 - val_accuracy: 0.9790 Epoch 39/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1183 - accuracy: 0.9531 - val_loss: 0.0647 - val_accuracy: 0.9790 Epoch 40/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1127 - accuracy: 0.9577 - val_loss: 0.0676 - val_accuracy: 0.9790 Epoch 41/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1092 - accuracy: 0.9671 - val_loss: 0.0603 - val_accuracy: 0.9790 Epoch 42/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1150 - accuracy: 0.9577 - val_loss: 0.0616 - val_accuracy: 0.9790 Epoch 43/100 7/7 [==============================] - 0s 6ms/step - loss: 0.0819 - accuracy: 0.9789 - val_loss: 0.0620 - val_accuracy: 0.9790 Epoch 44/100 7/7 [==============================] - 0s 5ms/step - loss: 0.0864 - accuracy: 0.9765 - val_loss: 0.0606 - val_accuracy: 0.9790 Epoch 45/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0922 - accuracy: 0.9671 - val_loss: 0.0625 - val_accuracy: 0.9790 Epoch 46/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0802 - accuracy: 0.9648 - val_loss: 0.0584 - val_accuracy: 0.9790 Epoch 47/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1113 - accuracy: 0.9624 - val_loss: 0.0574 - val_accuracy: 0.9790 Epoch 48/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1092 - accuracy: 0.9554 - val_loss: 0.0578 - val_accuracy: 0.9790 Epoch 49/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0921 - accuracy: 0.9742 - val_loss: 0.0616 - val_accuracy: 0.9790 Epoch 50/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0925 - accuracy: 0.9765 - val_loss: 0.0554 - val_accuracy: 0.9790 Epoch 51/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0931 - accuracy: 0.9648 - val_loss: 0.0561 - val_accuracy: 0.9790 Epoch 52/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1025 - accuracy: 0.9671 - val_loss: 0.0665 - val_accuracy: 0.9790 Epoch 53/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0977 - accuracy: 0.9695 - val_loss: 0.0560 - val_accuracy: 0.9790 Epoch 54/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1016 - accuracy: 0.9671 - val_loss: 0.0558 - val_accuracy: 0.9790 Epoch 55/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0844 - accuracy: 0.9742 - val_loss: 0.0650 - val_accuracy: 0.9790 Epoch 56/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0929 - accuracy: 0.9695 - val_loss: 0.0545 - val_accuracy: 0.9790 Epoch 57/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0744 - accuracy: 0.9812 - val_loss: 0.0539 - val_accuracy: 0.9790 Epoch 58/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0764 - accuracy: 0.9742 - val_loss: 0.0562 - val_accuracy: 0.9790 Epoch 59/100 7/7 [==============================] - 0s 4ms/step - loss: 0.1034 - accuracy: 0.9648 - val_loss: 0.0594 - val_accuracy: 0.9790 Epoch 60/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0789 - accuracy: 0.9695 - val_loss: 0.0529 - val_accuracy: 0.9790 Epoch 61/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0934 - accuracy: 0.9624 - val_loss: 0.0534 - val_accuracy: 0.9790 Epoch 62/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0804 - accuracy: 0.9695 - val_loss: 0.0601 - val_accuracy: 0.9790 Epoch 63/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0832 - accuracy: 0.9695 - val_loss: 0.0565 - val_accuracy: 0.9790 Epoch 64/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0862 - accuracy: 0.9718 - val_loss: 0.0531 - val_accuracy: 0.9790 Epoch 65/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0751 - accuracy: 0.9812 - val_loss: 0.0512 - val_accuracy: 0.9860 Epoch 66/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0724 - accuracy: 0.9812 - val_loss: 0.0530 - val_accuracy: 0.9790 Epoch 67/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0882 - accuracy: 0.9624 - val_loss: 0.0674 - val_accuracy: 0.9720 Epoch 68/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0784 - accuracy: 0.9765 - val_loss: 0.0497 - val_accuracy: 0.9860 Epoch 69/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0732 - accuracy: 0.9789 - val_loss: 0.0531 - val_accuracy: 0.9790 Epoch 70/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0799 - accuracy: 0.9718 - val_loss: 0.0531 - val_accuracy: 0.9790 Epoch 71/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0676 - accuracy: 0.9836 - val_loss: 0.0503 - val_accuracy: 0.9790 Epoch 72/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0647 - accuracy: 0.9789 - val_loss: 0.0522 - val_accuracy: 0.9790 Epoch 73/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0571 - accuracy: 0.9836 - val_loss: 0.0514 - val_accuracy: 0.9790 Epoch 74/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0656 - accuracy: 0.9812 - val_loss: 0.0516 - val_accuracy: 0.9790 Epoch 75/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0640 - accuracy: 0.9742 - val_loss: 0.0566 - val_accuracy: 0.9790 Epoch 76/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0654 - accuracy: 0.9742 - val_loss: 0.0535 - val_accuracy: 0.9790 Epoch 77/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0660 - accuracy: 0.9765 - val_loss: 0.0513 - val_accuracy: 0.9790 Epoch 78/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0576 - accuracy: 0.9812 - val_loss: 0.0516 - val_accuracy: 0.9790 Epoch 79/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0632 - accuracy: 0.9789 - val_loss: 0.0491 - val_accuracy: 0.9790 Epoch 80/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0784 - accuracy: 0.9695 - val_loss: 0.0507 - val_accuracy: 0.9790 Epoch 81/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0629 - accuracy: 0.9812 - val_loss: 0.0585 - val_accuracy: 0.9790 Epoch 82/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0709 - accuracy: 0.9789 - val_loss: 0.0541 - val_accuracy: 0.9790 Epoch 83/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0654 - accuracy: 0.9836 - val_loss: 0.0524 - val_accuracy: 0.9790 Epoch 84/100 7/7 [==============================] - 0s 3ms/step - loss: 0.0678 - accuracy: 0.9812 - val_loss: 0.0510 - val_accuracy: 0.9790 Epoch 85/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0843 - accuracy: 0.9648 - val_loss: 0.0505 - val_accuracy: 0.9790 Epoch 86/100 7/7 [==============================] - 0s 5ms/step - loss: 0.0645 - accuracy: 0.9836 - val_loss: 0.0518 - val_accuracy: 0.9790 Epoch 87/100 7/7 [==============================] - 0s 5ms/step - loss: 0.0567 - accuracy: 0.9859 - val_loss: 0.0529 - val_accuracy: 0.9790 Epoch 88/100 7/7 [==============================] - 0s 5ms/step - loss: 0.0613 - accuracy: 0.9765 - val_loss: 0.0561 - val_accuracy: 0.9790 Epoch 89/100 7/7 [==============================] - 0s 5ms/step - loss: 0.0730 - accuracy: 0.9671 - val_loss: 0.0555 - val_accuracy: 0.9790 Epoch 90/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0498 - accuracy: 0.9859 - val_loss: 0.0535 - val_accuracy: 0.9790 Epoch 91/100 7/7 [==============================] - 0s 7ms/step - loss: 0.0737 - accuracy: 0.9742 - val_loss: 0.0528 - val_accuracy: 0.9790 Epoch 92/100 7/7 [==============================] - 0s 8ms/step - loss: 0.0559 - accuracy: 0.9859 - val_loss: 0.0498 - val_accuracy: 0.9790 Epoch 93/100 7/7 [==============================] - 0s 7ms/step - loss: 0.0574 - accuracy: 0.9836 - val_loss: 0.0535 - val_accuracy: 0.9790 Epoch 94/100 7/7 [==============================] - 0s 6ms/step - loss: 0.0461 - accuracy: 0.9883 - val_loss: 0.0546 - val_accuracy: 0.9790 Epoch 95/100 7/7 [==============================] - 0s 8ms/step - loss: 0.0527 - accuracy: 0.9836 - val_loss: 0.0523 - val_accuracy: 0.9790 Epoch 96/100 7/7 [==============================] - 0s 6ms/step - loss: 0.0630 - accuracy: 0.9789 - val_loss: 0.0519 - val_accuracy: 0.9790 Epoch 97/100 7/7 [==============================] - 0s 7ms/step - loss: 0.0649 - accuracy: 0.9718 - val_loss: 0.0516 - val_accuracy: 0.9790 Epoch 98/100 7/7 [==============================] - 0s 9ms/step - loss: 0.0642 - accuracy: 0.9718 - val_loss: 0.0502 - val_accuracy: 0.9860 Epoch 99/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0622 - accuracy: 0.9812 - val_loss: 0.0544 - val_accuracy: 0.9790 Epoch 100/100 7/7 [==============================] - 0s 4ms/step - loss: 0.0695 - accuracy: 0.9765 - val_loss: 0.0535 - val_accuracy: 0.9790
Make a one-time donation
Make a monthly donation
Make a yearly donation
Choose an amount
Or enter a custom amount
Your contribution is appreciated.
Your contribution is appreciated.
Your contribution is appreciated.
DonateDonate monthlyDonate yearly